Genomic Technology Core
Committee members:
Dr. Chu, Hsiu-An (chair)
Dr. Chen, Pao-Yang
Dr. Wu, Chih-Hang
Dr. Wu, Ting-Ying
Dr. Yu, Tien-Shin
G-TeC Microarray and Sequencing Division :
Staff :
Shu-Jen Chou (Research Specialist)
Ai-Ping Chen (Research Assistant)
Contact Information:
02-27871072 (Office)
02-27871071 (Lab)
E.mail :
sjchou@gate.sinica.edu.tw
Location:
A222, Agricultural Technology Building
G-TeC DNA Analysis Division :
Staff :
Mei-Jane Fang (Research Assistant)
Ming-Ling Cheng (Research Assistant)
Contact Information :
02-27871028 (Office)
02-27871029 (Lab)
E.mail :
zofangmj@gate.sinica.edu.tw
Location :
Room 208,Institute of Plant and Microbial Biology
 Genomic Technology Core
Genomic Technology Core
The Genomic Technology Core (G-TeC) provides services related to gene expression (microarray, sequencing library preparation and quantitative PCR), genomic analysis (sequencing library preparation) related technologies. G-TeC actively develops new methods and assists IPMB researchers as well as additional users in Academia Sinica and Taiwan to taking full advantage of rapidly developing genome analysis technologies. G-TeC has two divisions which provide a complementary range of services
1.G-TeC Microarray and Sequencing Division
This division of G-TeC has a full suite of equipment for microarray experiments using Agilent/Exiqon DNA Microarrays (including Agilent 2100 Bioanalyzer, Agilent Microarray Scanner and Agilent hybridization oven) as well as analysis of microarray data (Genespring software).
In recent years, use of Next Generation Sequencing (NGS) technology has rapidly advanced. NGS technology has a range of applications in gene expression, small RNA and non-coding RNA analysis, ChIP sequencing, SNP detection and mapping as well as de novo genome sequencing. Preparation of sequencing libraries is key to obtaining reliable NGS results and has different requirements depending on application. G-TeC provides library preparation services for a range of NGS applications and can develop new library preparation protocols in response to user requests. We also have established contacts so that NGS libraries prepared in G-TeC can be passed on to NGS core in AS or commercial sequencing companies for analysis using the latest generation of NGS technology. The overall workflow provides high quality sequencing libraries and is both cost effective and convenient for users.
Current NGS library preparation services include:
► Illumina sequencing
- Genomic DNA library preps
- Stranded specific RNA-Seq library preps (poly-A enrichment or rRNA-depletion)
- miRNA-Seq library preps
- Mate Pair library preps
- Library preps from ChIP samples
- Parallel analysis of RNA end (PARE) library preps
- RiboSeq library preps
- PAT (polyA Tail) RNAseq library
- 16S/ITS amplicon library (New)
- Enzymatic Methyl-seq (EM-seq) library (New)
- Low input RNAseq library (>10 ug RNA)
- SMARTer-seq Ultra Low Input RNA library (>10 pg RNA) (New)
►PacBio long insert DNA sequencing library preparation.
►Oxford Nanopore Technology (ONT) library construction and sequencing
- Genomic DNA and native barcoding genomic DNA sequencing
- Direct RNA sequencing (New)
- Direct cDNA barcoding sequencing (New)
We also provide QX200 Droplet Digital PCR System, Agilent Bioanalyzer, Fragment Analyzer and Qubit Fluorometric Quantitation services.
►Fee schedules for IPMB user Non-IPMB user, please contact to Shu-Jen for detail information.
►Sample submission form (Bioanalyzer, Fragment Analyzer, Microarray and NGS library construction)Non-IPMB user, please contact to Shu-Jen for detail information.



2. G-TeC DNA Analysis Division
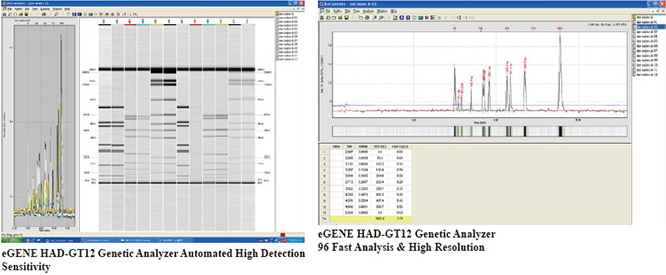
We provide routine DNA sequencing (using ABI 3730 DNA Analyzer) and Real-time quantitative PCR (Applied Biosystems (AB) QuantStudio 12K, 7500, and StepOne Plus real time PCR systems). The goal of this section of G-TeC is to provide cost effective solutions for routine DNA sequencing and Quantitative PCR including centralized purchasing of consumables and troubleshooting for quantitative PCR and DNA sequencing. The facility also maintains an eGENE HAD-GT12 Genetic Analyzer for common use.
Samples for routine DNA sequencing can be submitted daily to 11:00 A.M. The typical turnaround time from submission of DNA to availability of sequence is 1 day. Cost per sample is NT 80. Peak Trace: Post-run analysis of sequence traces by Peak Trace software can increase useable read length for sequences over 700-800 bp. Peak Trace is typically used for plasmid DNA templates. There is an additional charge of 3 NT/sample for Peak Trace analysis; however, this extra charge is only applied if Peak Trace successfully extends the useable read length. The QuantStudio 12K Flex Real-Time PCR System could complete the HRM experiment.
Scheduling of QPCR instrument usage is available through reservation.
►Sample submission form for DNA sequencing