Bioinformatics Core Lab

Committee Member
Associate Research Specialist
Research Assistant
- Yung-I Lin
Bioinformatics Core Facility was established in early 2007. The objective of the core is to apply bioinformatics expertise to generate interdisciplinary results. To do that, we provide the following types of services:
1. Consultation: Answering short questions on using bioinformatics software and statistics methods.
2. Computation: Executing user-specified software for specific biological research problems.
3. Research & Development: Customization of necessary bioinformatics tools, and organization of bioinformatics tools for stable operating procedures.
4. Online computation: An inhouse Galaxy system was depolyed to provide our working tools and essential tools, e.g. BLAST & MEME, with less computational limitation than most public online services. Please see our Core Lab area under Intranet for links to documents and services.
5.IPMB HPC: We help maintaining the HPC system owned by IPMB. Please refer the wiki page (intranet link) for more information
For IPMB members, please refer our Core Labs area under Intranet for links to workshop slides and inhouse resource locations.
Office hour
We will be at room R106 of the IPMB building for every Friday 1:30PM~2:30PM. This office hour will be suspended if other official events are scheduled. We also welcome people to contact us directly for discussions of bioinformatics related questions. For example, data processing using Excel, simple statistics analysis, suggestions on using public databases, or for a second opinion on bioinformatics analysis.
Task initiation
No specific forms are required to fill for initiating a service task. Initial conversation would be required for formalizing requests and turning them into computationally doable form. Please write us an email for making an appointment. It is appreciated if the context of the request could be addressed first.
Charges
The service charge is 1100NTD/working hour for IPMB members. Here are two examples of working hours for Arabidopsis thaliana according to our recent records:
1. RNAseq task: 40M read-sample x 15 will take about 3 working days. The output includes differentially expressed gene discovery and alternative-splicing comparisons.
2. DNAseq task: 50M read-sample x 9 will take about 2.5 working days for SNP/InDel calling and annotation.
Service experiences
Service categories that we have experienced. Our services are including but not limited to the following aspects:
DNAseq data
Sequence assembly
SNP/InDel detection
T-DNA insertion site detection
Tandem duplication detection
Genome rearrangement detection
RNAseq data
Expression level computation
Sample-sensitive alternative splicing events
Alternative polyadenylation detection
Gene fusion detection
scRNAseq data processing
Other NGS analyses
SmallRNA expression level computation
SmallRNA target prediction
ChIPseq data processing
Systems biology
Co-expression clustering
PPI network computation
Sequence motif searching
GO enrichment computation
Automatic GO annotation
Miscellaneous
Enzyme kinetics computation
Computation of isothermal tiling array probes
iTRAQ data processing
Protein structure and interaction prediction
Open source projects
This bioinformatics core facility is also maintaining the following three open source projects.
1. MACCU (Multi-Array Correlation Computation Utility, https://github.com/wdlingit/maccu ):Computation of co-expression networks and comparisons between networks which help finding tissue-specific co-expression modules.
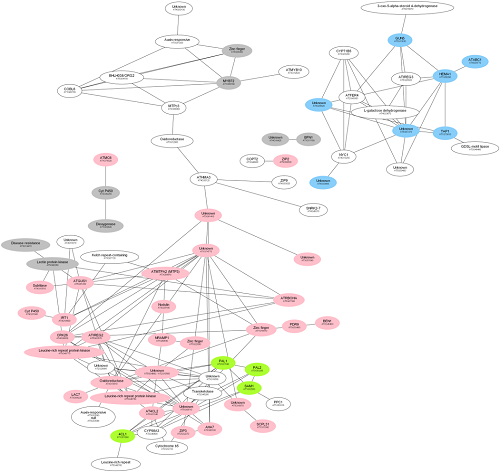
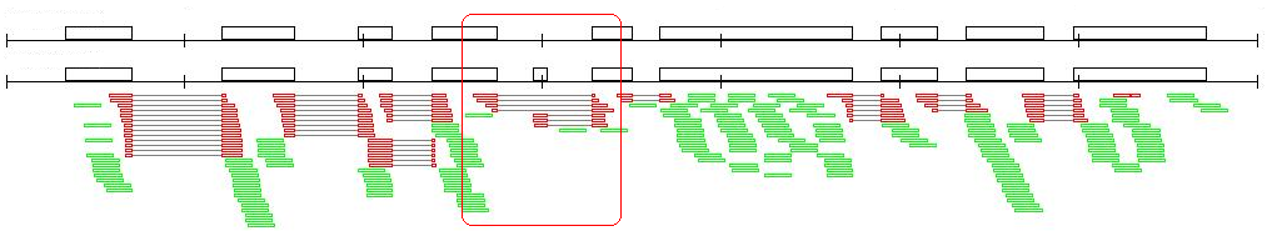
2. RackJ (Read Analysis & Comparison Kit in Java, http://rackj.sourceforge.net/ ):In addition to computation of RPKM values of RNAseq data, this toolkit computes read numbers for every exon, intron, and splicing junction, thus enables finding of sample-sensitive alternative-splicing events.
3. GOBU (Gene Ontology Browsing Utility, https://gobu.sourceforge.io/ ):A user-friendly graphical interface program for GO data manipulation, including fast computation modules for enrichment analysis.
Selected Publications
Huang CK, Lin WD, Wu SH (2022) An improved repertoire of splicing variants and their potential roles in Arabidopsis photomorphogenic development. Genome Biol. 9;23(1):50.
Hsieh EJ, Lin WD, Schmidt W (2022) Genomically Hardwired Regulation of Gene Activity Orchestrates Cellular Iron Homeostasis in Arabidopsis. RNA Biol. 19(1):143-161.
Kanno T, Venhuizen P, Wen TN, Lin WD, Chiou P, Kalyna M, Matzke AJM, Matzke M (2018) PRP4KA, a Putative Spliceosomal Protein Kinase, Is Important for Alternative Splicing and Development in Arabidopsis thaliana. Genetics. 210(4):1267-1285.
Salazar-Henao JE, Lin WD, Schmidt W (2016) Discriminative gene co-expression network analysis uncovers novel modules involved in the formation of phosphate deficiency-induced root hairs in Arabidopsis. Sci Rep 6:26820.
Kanno T, Lin WD, Fu JL, Wu MT, Yang HW, Lin SS, Matzke AJ, Matzke M (2016 Identification of Coilin Mutants in a Screen for Enhanced Expression of an Alternatively Spliced GFP Reporter Gene in Arabidopsis thaliana. Genetics 203(4):1709-20.
Sasaki T, Kanno T, Liang SC, Chen PY, Liao WW, Lin WD, Matzke AJ, Matzke M (2015) An Rtf2 domain-containing protein influences pre-mRNA splicing and is
essential for embryonic development in Arabidopsis thaliana. Genetics 200(2):523-35.
Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, López-Millán AF, and Schmidt W. (2013) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by arabidopsis and Medicago truncatula. Plant Physiology. 162:1473-85.
Li W, Lin WD, Ray P, Lan P, and Schmidt W. (2013) Genome-wide detection of condition-sensitive alternative splicing in Arabidopsis roots. Plant Physiology. 162:1750-63.
Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, and Wu SH.. (2013) Translational landscape of photomorphogenic Arabidopsis. Plant Cell. 25:3699-710.
Lan P, Li WF, Lin WD, Santi S, and Schmidt W. (2013) Mapping gene activity of Arabidopsis root hairs. Genome Biology. Genome Biology. 14:R67.
Chen YT, Shen CH, Lin WD, Chu HA, Huang BL, Kuo CI, Yeh KW, Huang LC, and Chang IF. (2013) Small RNAs of Sequoia sempervirens during rejuvenation and phase change. Plant Biology. 15:27-36.
Sharma S, Lin WD, Villamor JG, and Verslues PE. (2013) Divergent low water potential response in Arabidopsis thaliana accessions Landsberg erecta and Shahdara. Plant Cell & Environment 36: 994-1008.
Chen YC, Chen YC, Lin WD, Hsiao CD, Chiu HW, and Ho JM. (2012) Bio301: A Web-based EST annotation pipeline that facilitates functional comparison studies. ISRN Bioinformatics. (doi:10.5402/2012/139842)
Lin WD, Liao YY, Yang TJ, Pan CY, Buckhout TJ, and Schmidt W. (2011) Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiology. 155:1383-402.