
Chen, Pao-Yang (陳柏仰)
Research Fellow
- Ph.D., Statistics, Oxford University, UK
- Genome-wide DNA methylation, Epigenomics and Bioinformatics
- paoyang@gate.sinica.edu.tw
- paoyang@as.edu.tw
- +886-2-2787-1139 (Lab: R316)
- Lab Website
- Academia Sinica Archive
- ORCID
- Web of Science (WOS)
- Google Scholar
The focus of our research is on the development of both experimental and computational methods to interpret genomic and epigenomic data from plants, animals, fungi and human. By integrating data produce from the next-generation sequencing technologies, we would like to implement novel analytical strategies to solve problems in genetics and molecular biology.
Next generation genetics:
Next-generation or massively parallel DNA sequencing technologies have the potential to markedly accelerate genetics research. In our lab we perform DNA-seq for whole genome assembly, mRNA-seq for transcriptome studies, and BS-seq for DNA methylation profiling (WGBS and RRBS). Our goal is to integrate both experiments and statistical methods and software to address biologically relevant questions.
DNA methylation in plants:
Cytosine DNA methylation is an epigenetic modification of DNA that is usually associated with the stable and heritable repression of transcription. We would like to study DNA methylation in plants as their small genome sizes allow them to be sequenced economically. Plants are also easy to sample and tend to be amenable to genetic manipulations; the annotations of model plants are also well developed. Plant methylomes can provide important information for clarifying the mechanisms of action and characteristics of DNA methyltransferase enzymes, as well as contributing to a better understanding of the evolution of gene promoters and other regulatory sequences. Currently, we are investigating the genome wide DNA methylation pattern in maize meiocytes, rice transformation, and the stress responses in plants.
DNA methylation landscape in plants, animals, and human:
The genomes of many animals, plants and fungi are tagged by methylation of DNA cytosine. To understand the biological significance of this epigenetic mark it is essential to know where in the genome it is located. Our lab has several collaboration projects with UCLA to study the DNA methylation patterns in mammalian germ cells. For example, we are investigating the genome wide DNA methylation pattern on human prenatal germ cells. In addition, we are examining the impact of IVF (in vitro fertilization) on human and mice by studying the methylation status in fetus. New techniques are making it easier to map DNA methylation patterns on a large scale and the results have already provided surprises. We aimed to develop tools to understand the impact of this highly dynamic DNA methylation pattern and its function.
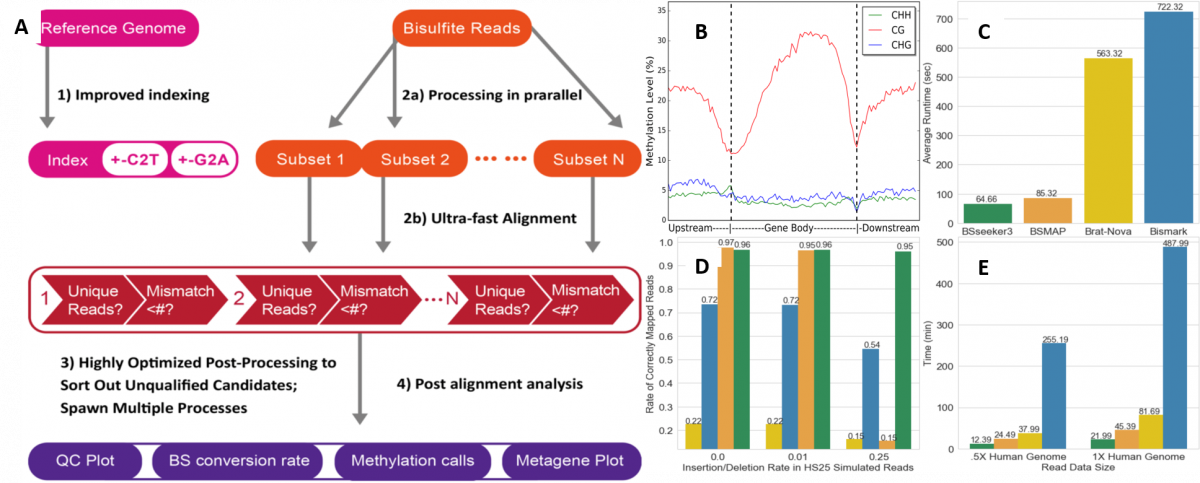
Summary of BS-Seeker3 pipeline and performance.
- (A)Schematic flow chart of BS Seeker 3 with improved indexing, data processing, fast alignment and post-alignment analyses
- (B)Metaplot of Methylation level: This metaplot presents the average methylation level distribution within a user-specified genomic structure (e.g., coding genes) inArabiodopsis thalania. CG denotes a CpG dinucleotide, CHG denotes a cytosine next to a H where H stands for A, C, or T.and then a guanine, CHH denotes a cytosine next to two H bases
- (C)Average user runtime of the four aligners on 10M simulated HiSeq 2500 Arabidopsis reads.
- (D)Percentage of the 10M simulated HiSeq2500 reads that were mapped correctly across various reads complexity level.
- (E)Average runtime of four aligners on directional BS-seq reads from real human data.
- Pei-Yu Lin, Guan-Jun Lin, Kuan-Lin Chen, Shiang-Chin Huang, Pao-Yang Chen¶ (2025) Computational Workflow for Genome-Wide DNA Methylation Profiling and Differential Methylation Analysis. Bio-protocol, 15(21): e5506.
- Fei-Man Hsu, Matteo Pellegrini¶, Pao-Yang Chen¶(2025) Library Preparation for Genome-Wide DNA Methylation Profiling. Bio-protocol, 15, 5(21): e5488.
- Pei-Yu Lin, Shiang-Chin Huang, Kuan-Lin Chen, Yu-Chun Huang, Chia-Yu Liao, Guan-Jun Lin, HueyTyng Lee, and Pao-Yang Chen¶ (2025) Analysing Protein Complexes in Plant Science: Insights and limitation with AlphaFold 3. Botanical Studies, 66,14.
- Kuan-Ting Hsin, HueyTyng Lee, Yu-Chun Huang, Guan-Jun Lin, Pei-Yu Lin, Ying-Chung Jimmy Lin, and Pao-Yang Chen¶ (2025) Lignocellulose Degradation in Bacteria and Fungi: Cellulosomes and Industrial Relevance. Frontiers in Microbiology, 16, 1583746.
- Debamalya Chatterjee, Ziru Zhang, Pei-Yu Lin, Po-Hao Wang, Gurpreet K Sidhu, Neela H Yennawar, Jo-Wei Allison Hsieh, Pao-Yang Chen, Rentao Song, Blake Meyers and Surinder Chopra¶ (2025) Maize unstable factor for orange1 encodes a nuclear ptotein that affects redox accumulation during kernel development. The Plant Cell, 37(1): koae301.
- Chih-Hung Hsieh, Ya-Ting Sabrina Chang, Ming-Ren Yen, Jo-Wei Allison Hsieh and Pao-Yang Chen¶ (2024) Predicting protein synergistic effect in Arabidopsis using epigenome profiling. Nature Communications, 15(1): 9160.
- Jo-Wei Allison Hsieh, Ming-Ren Yen, Fu-Yu Hung, Keqiang Wu¶ and Pao-Yang Chen¶ (2024) Epigenetic factors direct synergistic and antagonistic regulation of transposable elements in Arabidopsis. Plant Physiology, kiae392, https://doi.org/10.1093/plphys/kiae392.
- Ti-Wen Lu, Wen-Huei Chen, Pao-Yang Chen, Yu-Chen Shu, Hong-Hwa Chen¶ (2024) Perturbation of periodic spot-generation balance leads to diversified pigmentation patterning of harlequin Phalaenopsis orchids: in silico prediction. BMC Plant Biology, 24: 681.
- Jo-Wei Allison Hsieh*, Pei-Yu Lin*, Chi-Ting Wang, Yi-Jing Lee, Pearl Chang, Rita Jui-Hsien Lu, Pao-Yang Chen¶ and Chung-Ju Rachel Wang¶ (2024) Establishing an optimized ATAC-seq protocol for the maize. Frontiers in Plant Science,15:1370618.
- Pei-Yu Lin, Kuan-Lin Chen, Guan-Jun Lin, Shiang-Chin Huang and Pao-Yang Chen¶ (2024) Bioinformatics analysis of DNA methylation. Methods in Molecular Biology, DNA methylation edition, Springer Nature, accepted.
- Fei-Man Hsu, Pao-Yang Chen and Matteo Pellegrini¶ (2024) Whole genome bisulfite sequencing. Methods in Molecular Biology, DNA methylation edition, Springer Nature, accepted.
- Chiao-Yu Lyra Sheu*, Yu-Chun Huang*, Pei-Yu Lin, Guan-Jun Lin and Pao-Yang Chen¶ (2024) Chapter 3: Bioinformatics of epigenetic data generated from next-generation sequencing. Epigenetics in Human Disease, 3rd edition, Elsevier,1: 37-82.
- Pei-Yu Lin*, Ya-Ting Sabrina Chang*, Yu-Chun Huang and Pao-Yang Chen¶ (2023) Estimating genome-wide DNA methylation heterogeneity with methylation patterns. Epigenetics & Chromatin, 16(1): 44.
- HueyTyng Lee, Pei-Yu Lin and Pao-Yang Chen¶ (2023) There’s more to it: uncovering genomewide DNA methylation heterogeneity. Epigenomics 15(13): 687-691.
- Jo-Wei Allison Hsieh, Pearl Chang, Lin-Yun Kuang, Yue-Ie Hsing, Pao-Yang Chen¶ (2023) Rice transformation treatments leave specific epigenome changes beyond tissue culture. Plant Physiology, 193(2): 1297-1312.
- Rita Jui-Hsien Lu*, Pei-Yu Lin*, Ming-Ren Yen*, Bing-Heng Wu and Pao-Yang Chen (2023) MethylC-analyzer: a comprehensive downstream pipeline for the analysis of genome-wide DNA methylation. Botanical Studies 64(1):1.
- Pierre Bourguet, Ramesh Yelagandula, Taiko Kim To, Akihisa Osakabe, Archana Alishe, Rita Jui-Hsien Lu, Tetsuji Kakutani, Pao-Yang Chen¶ and Frédéric Berger¶ (2022) The histone variant H2A.W cooperates with chromatin modifications and linker histone H1 to maintain transcriptional silencing of transposons in Arabidopsis. bioRxiv https://doi.org/10.1101/2022.05.31.493688
- Yi-Tsung Tu, Chia-Yang Chen, Yi-Sui Huang, Chung-Han Chang, Ming-Ren Yen, Jo-Wei Allison Hsieh, Pao-Yang Chen¶ and Keqiang Wu¶ (2022) HISTONE DEACETYLASE 15 and MOS4-associated complex subunits 3A/3B coregulate intron retention of ABA-responsive genes. Plant Physiology 190(1): 882-897.
- Matthew Lowe, Ming-Ren Yen, Fei-Man Hsu, Linzi Hosohama, Zhongxun Hu, Tsotne Chitiashvili, Timothy Hunt, Isaac Gorgy, Matthew Bernard, S Wamaitha, Pao-Yang Chen¶ and Amander T Clark¶ (2022) EED is required for Mouse Primordial Germ Cell Differentiation in the Embryonic Gonad. Developmental Cell 57(12): 1482-1495.e5.
- Yu-Wen Liu, Shin-Huei Fu, Ming-Wei Chien, Chao-Yuan Hsu, Ming-Hong Lin, Jia-Ling Dong, Rita Jui-Hsien Lu, Yi-Jing Lee, Pao-Yang Chen, Chih-Hung Wang, and Huey-Kang Sytwu (2022) Blimp-1 moulds the epigenetic architecture of IL-21-mediated autoimmune diseases through an autoregulatory circuit. JCI Insight https://doi.org/10.1172/jci.insight.151614.
- Feng-Chih Kuo¶, Yu-Chun Huang, Ming-Ren Yen, Chien-Hsing Lee, Kuo-Feng Hsu, Hsiang-Yu Yang, Li-Wei Wu, Chieh-Hua Lu, Yu-Juei Hsu and Pao-Yang Chen¶ (2022) Aberrant overexpression of HOTAIR inhibits abdominal adipogenesis through remodelling of genome-wide DNA methylation and transcription. Molecular Metabolism https://doi.org/10.1016/j.molmet.2022.101473.
- Ya-Ting Sabrina Chang, Ming-Ren Yen and Pao-Yang Chen (2022) BSImp: imputing partially observed methylation patterns for evaluating methylation heterogeneity. Frontiers in Bioinformatics https://doi.org/10.3389/fbinf.2022.815289.
- Chih-Yi Yang*, Jui-Hsien Lu*, Ming-Kang Lee*, Shih-Hsiang Felix Hsiao, Ya-Ping Yen, Chun-Chun Cheng, Pu-Sheng Hsu, Yi-Tzang Tsai, Shih-Kuo Chen, I-Hsuan Liu, Pao-Yang Chen¶ and Shau-Ping Lin¶ (2021) Transcriptome Analysis of Dnmt3l Knock-Out Mice Derived Multipotent Mesenchymal Stem/Stromal Cells During Osteogenic Differentiation. Frontiers in Cell and Developmental Biology, section Stem Cell Research 9: 615098.
- Yu-Shin Nai*, Yu-Chun Huang*, Ming-Ren Yen and Pao-Yang Chen (2021) Diversity of fungal DNA methyltransferases and their association with DNA methylation patterns. Frontiers in Microbiology, section Evolutionary and Genomic Microbiology 11: 616922.
- Rita Jui-Hsein Lu*, Yen-Ting Liu*, Chih Wei Huang*, Ming-Ren Yen, Chung-Yen Lin and Pao-Yang Chen (2021) ATACgraph: profiling genome wide chromatin accessibility from ATAC-seq. Frontiers in Genetics 11: 618478.
- Huei-Mien Ke¶, Hsin-Han Lee, Chan-Yi Ivy Lin, Yu-Ching Liu, Min R. Lu, Jo-Wei Allison Hsieh, Chiung-Chih Chang, Pei-Hsuan Wu, Meiyeh Jade Lu, Jeng-Yi Li, Gaus Shang, Rita Jui-Hsien Lu, László G. Nagy, Pao-Yang Chen, Hsiao-Wei Kao and Isheng Jason Tsai¶ (2020) Mycenagenomes resolve the evolution of fungal bioluminescence. Proceedings of the National Academy of Sciences 117(49): 31267-31277.
- Fu-Yu Hung, Chen Chen, Ming-Ren Yen, Jo-Wei Hsieh, Chenlong Li, Yuan-Hsin Shih, Fang-Fang Chen, Pao-Yang Chen¶, Yuhai Cui¶ and Keqiang Wu¶ (2020) The expression of long noncoding RNAs is associated with H3Ac and H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex in Arabidopsis. NAR Genomics and Bioinformatics 2(3): lqaa066.
- Hong Zhu, Pao-Yang Chen, Silin Zhong, Christopher Dardick, Ann Callahan, Yong-Qiang An, Steve van Nocker, Yingzhen Yang, Gan-Yuan Zhong, Albert Abbott and Zongrang Liu (2020) Thermal-responsive genetic and epigenetic regulation of DAM cluster controlling dormancy and chilling requirement in peach floral buds. Horticulture Research 7: 114.
- Jo-Wei Allison Hsieh, Ming-Ren Yen and Pao-Yang Chen (2020) Epigenomic regulation of OTU5 in Arabidopsis thaliana. Genomics S0888-7543(20)30008-2.
- Chin-Sheng Teng, Bing-Heng Wu, Ming-Ren Yen and Pao-Yang Chen (2020) MethGET: web-based bioinformatics software for correlating genome-wide DNA methylation and gene expression. BMC Genomics 21(1): 375.
- Sheng-Yao Su, I-Hsuan Lu, Wen-Chih Cheng, Wei-Chun Chung, Pao-Yang Chen, Jan-Ming Ho, Shu-Hwa Chen¶ and Chung-Yen Lin¶ (2020) EpiMOLAS: an intuitive web-based framework for genome-wide DNA methylation analysis. BMC Genomics 21(Suppl 3): 163.
- Emilie debladis, Tzuu-Fen Lee, Yan-Jiun Huang, Jui-Hsien Lu, Sandra Mathioni, Marie-Christine Carpentier, Christel Llauro, Davy Pierron, Delphine Mieulet, Emmanuel Guiderdoni, Pao-Yang Chen, Blake Meyers, Olivier Panaud and Eric Lasserre (2020) Construction and characterization of a knock-down RNA interference line of OsNRPD1 in rice (Oryza sativa ssp. Japonica cv. Nipponbare). Philosophical Transactions of the Royal Society B 375: 20190338.
- Jie Yang*, Lianyu Yuan*, Ming-Ren Yen*, Tao Peng, Dachuan Gu, Rujun Ji, Songguang Yang, Yuhai Cui, Pao-Yang Chen, Keqiang Wu¶ and Xuncheng Liu¶ (2020) SWI3B and HDA6 interact and are required for transposon silencing in Arabidopsis. The Plant Journal 102(4): 809-822.
- You-Yuan Pang*, Jui-Hsien Lu* and Pao-Yang Chen (2019) Behavioral epigenetics: perspectives based on experience-dependent epigenetic inheritance. Epigenomes 3(3): 18.
- Phuong Nguyen Tran, Ming-Ren Yen, Chen-Yu Chiang, Hsiao-Ching Lin¶ and Pao-Yang Chen¶ (2019) Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Applied Microbiology and Biotechnology (103) 8: 3277-3287.
- Ta-Ching Chen, Pin-Yi She, Dong-Feng Chen, Jui-Hsien Lu, Chang-Hao Yang, Ding-Siang Huang, Pao-Yang Chen, Chen-Yu Lu, Kin-Sang Cho, Hsin-Fu Chen¶ and Wei-Fang Su¶ (2019) Polybenzyl glutamate biocompatible scaffold promotes the efficiency of retinal differentiation toward retinal ganglion cell lineage from human induced pluripotent stem cells. International Journal of Molecular Sciences 20(1): 178.
- Jie Hou*, Xiaowen Shi*, Chen Chen*, Md. Soliman Islam*, Adam F. Johnson*, Tatsuo Kanno, Bruno Huettel, Ming-Ren Yen, Fei-Man Hsu, Tieming Ji, Pao-Yang Chen, Marjori Matzke¶, Antonius J.M. Matzke¶, Jianlin Cheng and James A. Birchler¶ (2018) Global impacts of chromosomal imbalance on gene expression in Arabidopsis and other taxa. Proceedings of the National Academy of Sciences 115(48): E11321-E11330.
- Fei-Man Hsu*, Moloya Gohain*, Archana Allishe, Yan-Jiun Huang, Jo-Ling Liao, Lin-Yun Kuang and Pao-Yang Chen(2018) Dynamics of the methylome and transcriptome during the regeneration of rice. Epigenomes 2(3): 14.
- Yu-Yuan Huang, Yan-Jiun Huang and Pao-Yang Chen (2018) BS-Seeker3: ultrafast pipeline for bisulfite sequencing. BMC Bioinformatics 19: 111.
- Fei-Man Hsu, Chung-Ju Rachel Wang¶ and Pao-Yang Chen¶ (2018) Reduced representation bisulfite sequencing in maize. Bio-protocol 8(6): e2778.
- Pearl Chang, Yu-Fang Tseng, Pao-Yang Chen¶ and Chung-Ju Rachel Wang¶ (2018) Using flow cytometry to isolate maize meiocytes for next generation sequencing: a time and labor efficient method. Current Protocols in Plant Biology 3(2): e20068.
- Fei-Man Hsu, Moloya Gohain, Pearl Chang, Jui-Hsien Lu and Pao-Yang Chen (2018) Bioinformatics of epigenetic data generated from next-generation sequencing. Epigenetics in Human Disease, 2nd edition, Elsevier6 in Translational Epigenetics 65-106.
- Pearl Chang, Moloya Gohain, Ming-Ren Yen and Pao-Yang Chen (2018) Computational methods for assessing chromatin hierarchy. Computational and Structural Biotechnology Journal 16: 43-53.
- Yu Tao*, Ming-Ren Yen*, Tsotne Chitiashvili, Haruko Nakano, Rachel Kim, Linzi Hosohama, Yao-Chang Tan, Atsushi Nakano, Pao-Yang Chen¶ and Amander T. Clark¶ (2017) TRIM28-regulated transposon repression is required for human germline competency and not primed or naïve human pluripotency. Stem Cell Reports 10(1): 243-256.
- Jer-Young Lin*, Brandon H. Le*, Min Chen*, Kelli F. Henry, Jungim Hur, Tzung-Fu Hsieh, Pao-Yang Chen, Julie M. Pelletier, Matteo Pellegrini, Robert L. Fischer, John J. Harada and Robert B. Goldberg (2017) Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proceedings of the National Academy of Sciences 114(45): E9730-E9739.
- Ming-Ren Yen, Der-Fen Suen, Fei-Man Hsu, Yi-Hsiu Tsai, Hong-Yong Fu, Wolfgang Schmidt¶ and Pao-Yang Chen¶ (2017) Deubiquitinating enzyme OTU5 contributes to DNA methylation patterns and is critical for phosphate nutrition signals. Plant Physiology 175 (4): 1826-1838.
- Ta-Ching Chen*, Chao-Yuan Yeh*, Chao-Wen Lin, Chung-May Yang, Chang-Hao Yang, I-Hung Lin, Pao-Yang Chen, Jung-Yu Cheng and Fung-Rong Hu (2017) Vascular hypoperfusion in acute optic neuritis is a potentially new neurovascular model for demyelinating diseases. PLoS ONE 12(9): e0184927.
- Huei-Mei Hsieh, Mei-Chu Chung, Pao-Yang Chen, Fei-Man Hsu, Wen-Wei Liao, Ai-Ning Sung, Chun-Ru Lin, Chung-Ju Wang, Yu-Hsin Kao, Mei-Jane Fang, Chi-Yung Lai, Chieh-Chen Huang, Jyh-Ching Chou, Wen-Neng Chou, Bill Chia-Han Chang and Yu-Ming Ju (2017) A termite symbiotic mushroom maximizing sexual activity at growing tips of vegetative hyphae. Botanical Studies 58: 39.
- Fei-Man Hsu, Ming-Ren Yen, Chi-Ting Wang, Chien-Yu Lin, Chung-Ju Rachel Wang¶ and Pao-Yang Chen¶ (2017) Optimized reduced representation bisulfite sequencing reveals tissue-specific mCHH islands in maize. Epigenetics & Chromatin 10 (1): 42.
- Pao-Yang Chen*, Alison Chu*, Wen-Wei Liao, Liudmilla Rubbi, Carla Janzen, Fei-Man Hsu, Amit Ganguly, Shanthie Thamotharan, Matteo Pelligrini and Sherin Devaskar (2017) Prenatal growth patterns and birthweight are associated with differential DNA methylation and gene expression of cardiometabolic risk genes in human placentas. Reproductive Sciences 25(4): 523-539.
- Meng-Ying Wu, Chia-Yeh Lin, Hsin-Yi Tseng, Fei-Man Hsu, Pao-Yang Chen and Cheng-Fu Kao (2017) H2B ubiquitylation and the histone chaperone Asf1 cooperatively mediate the formation and maintenance of heterochromatin silencing. Nucleic Acids Research 45(14): 8225-8238.
- Fei-Man Hsu and Pao-Yang Chen¶ (2017) Game theory in epigenetic reprogramming: Comment on: Epigenetic game theory: How to compute the epigenetic control of maternal-to-zygotic transition by Qian Wang et al. Physics of Life Reviews 20: 143 (Invited commentary).
- Sheng-Yao Su, Shu-Hwa Chen, I-Hsuan Lu, Yih-Shien Chiang, Yu-Bin Wang, Pao-Yang Chen¶ and Chung-Yen Lin¶ (2016) TEA: the epigenome platform for Arabidopsis methylome study. BMC Genomics 17(13): 3326.
- Joseph Hargan-Calvopina, Sara Taylor, Helene Cook, Zhongxun Hu, Serena A. Lee, Ming-Ren Yen, Yih-Shien Chiang, Pao-Yang Chen and Amander T. Clark (2016) Stage-specific demethylation in primordial germ cells safeguards against precocious differentiation. Developmental Cell 39(1): 75-86.
- Wai-Shin Yong, Fei-Man Hsu and Pao-Yang Chen¶ (2016) Profiling genome-wide DNA methylation. Epigenetics & Chromatin 9: 26.
- Colin Flinders, Larry Lam, Liudmilla Rubbi, Roberto Ferrari, Sorel Fitz-Gibbon, Pao-Yang Chen, Michael Thompson, Heather Christofk, David B Agus, Daniel Ruderman, Parag Mallick and Matteo Pellegrini (2016) Epigenetic changes mediated by polycomb repressive complex 2 and E2a are associated with drug resistance in a mouse model of lymphoma. Genome Medicine 8(1): 54.
- Fei-Man Hsu, Wai-Shin Yong and Pao-Yang Chen¶ (2016) Views on profiling genomewide DNA methylation and crop methylomes Crop, Environment & Bioinformatics (in Chinese), 作物、環境與生物資訊 13: 39–51.
- Wen-Wei Liao, Ming-Ren Yen, Evaline Ju, Fei-Man Hsu, Larry Lam and Pao-Yang Chen¶ (2015) MethGO: a comprehensive tool for analyzing whole genome bisulfite sequencing data. BMC Genomics 16(Suppl 12): S11.
- Fei-Man Hsu, Amander T Clark and Pao-Yang Chen¶ (2015) Epigenetic reprogramming in the mammalian germline. Oncotarget 6 (34): 35151 (Invited review)
- Sofia Gkountela, Kelvin Zhang, Tiasha Shafiq, Joseph Hargan-Calvopina, Pao-Yang Chen¶ and Amander T Clark¶ (2015) DNA demethylation dynamics in the human prenatal germline. Cell 161: 1425–1436.
- Taku Sasaki, Tatsuo Kanno, Shih-Chieh Liang, Pao-Yang Chen, Wen-Wei Liao, Wen-Dar Lin, Antonius J.M. Matzke and Marjori Matzke (2015) An Rtf2 domain-containing protein influences pre-mRNA splicing and is essential for embryonic development in Arabidopsis thaliana. Genetics 200(2): 523.
- Pao-Yang Chen*, Barbara Montanini*, Wen-Wei Liao, Marco Morselli, Artur Jaroszewicz, David Lopez, Simone Ottonello and Matteo Pellegrini (2014) A comprehensive resource of genomic, epigenomic and transcriptomic sequencing data for the black truffle Tuber melanosporum. GigaScience 3(1): 25.
- Barbara Montanini*, Pao-Yang Chen*, Marco Morselli, Artur Jaroszewicz, David Lopez, Francis Martin, Simone Ottonello and Matteo Pellegrini (2014) Non-exhaustive DNA methylation-mediated transposon silencing in the black truffle genome, a complex fungal genome with massive repeat element content. Genome Biology 15(8): 411.
- Negar M Ghahramani, Tuck C Ngun, Pao-Yang Chen, Yuan Tian, Sangitha Krishnan, Stephanie Muir, Liudmilla Rubbi, Arthur P Arnold, Geert J de Vries, Nancy G Forger, Matteo Pellegrini and Eric Vilain (2014) The effects of perinatal testosterone exposure on the DNA methylome of the mouse brain are late-emerging. Biology of Sex Differences 5(1): 8.
- Taku Sasaki, Tzuu-fen Lee, Wen-Wei Liao, Ulf Naumann, Jo-Ling Liao, Changho Eun, Ya-Yi Huang, Jason L. Fu, Pao-Yang Chen, Blake C. Meyers, Antonius J.M. Matzke and Marjori Matzke (2014) Distinct and concurrent pathways of Pol II and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana. The Plant Journal 79(1): 127-138.
- Weilong Guo, Petko Fiziev, Weihong Yan, Shawn Cokus, Xueguang Sun, Michael Q Zhang, Pao-Yang Chen¶ and Matteo Pellegrini¶ (2013) BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14(1): 774 (co-corresponding authors)
- Pao-Yang Chen, Amit Ganguly, Liudmilla Rubbi, Luz D. Orozco, Marco Morselli, Davin Ashraf, Artur Jaroszewicz, Suhua Feng, Steve E. Jacobsen, Atsushi Nakano, Sherin U. Devaskar and Matteo Pellegrini (2013) Intra-uterine calorie restriction affects placental DNA methylation and gene expression. Physiological Genomics 45(14): 565-576.
- John J. Vincent, Yun Huang, Pao-Yang Chen, Suhua Feng, Joseph H. Calvopin, Kevin Nee, Serena A. Lee, Thuc Le, Alexander J. Yoon, Kym Faull, Guoping Fan, Anjana Rao, Steven E. Jacobsen, Matteo Pellegrini, and Amander T. Clark (2013) Stage-specific roles for Tet1 and Tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell 12(4): 470-8.
Domestic
- 2021: Professor Chu-Yung Lin Plant Biology Innovative Research Award, CY Lin Foundation for Plant Science and Education
- 2013: Recruiting International Outstanding Junior Scholar Award, Foundation for the Advancement of Outstanding Scholarship (FAOS)
