生物資訊核心實驗室

現任委員 Committee Member
研究副技師 Associate Research Specialist
助理 Research Assistant
- 林詠儀 Yung-I Lin
生物資訊核心實驗室設立於2007年初,設立目的為生物資訊專業知識的溝通管道,從而協助研究人員進行跨領域研究。目前我們提供以下形式的服務:
1.諮商:生物資訊軟體及統計方法的使用
2.計算:針對使用者的特定問題使用指定軟體進行運算
3.研發:客製化並整合不同生物資訊程式,使之穩定運算
4.線上運算:提供內部使用的Galaxy系統,內容有生物資訊核心實驗室正在使用的工具介面,以及其他必要工具(如BLAST及MEME)。請參考內部系統核心實驗室專區的網站及說明文件連結。
5.高效能運算:我們協助維護植微所內部高效能運算叢集,請參考相關說明網頁(內部系統連結)以取得更多資訊。
植微所同仁請參考內部系統核心實驗室專區的文件,內有過往投影片及相關所內資源連結。
諮詢時間
每周五下午一點半到兩點半,我們會在植微大樓R106會議室。如遇所方有其他活動則暫停。其他時間歡迎同仁直接與我們聯繫。歡迎各位同仁前來討論任何生物資訊相關問題。舉凡如何使用Excel處理資料,簡易統計分析,如何使用常用的公開資料庫,或者是生物資訊分析的討論。
服務流程
不需要填寫任何表格。我們需要初步的討論以確定需求並將問題轉換成可解決的形式,請以電子郵件和我們聯絡約定時間討論。歡迎同時在信件中提供問題的相關資訊。
收費標準
中研院植微所成員收費為每小時1100元。以下為兩例阿拉伯芥研究課題的收費計時紀錄:
1. RNAseq:15個40M read樣本,耗時約三個工作天。結果包括differentially expressed gene discovery及alternative-splicing comparisons。
2. DNAseq:9個50M樣本,耗時約兩個半工作天。結果包括SNP/InDel calling及其註解。
服務紀錄
DNAseq data
Sequence assembly
SNP/InDel detection
T-DNA insertion site detection
Tandem duplication detection
Genome rearrangement detection
RNAseq data
Expression level computation
Sample-sensitive alternative splicing events
Alternative polyadenylation detection
Gene fusion detection
scRNAseq data processing
Other NGS analyses
SmallRNA expression level computation
SmallRNA target prediction
ChIPseq data processing
Systems biology
Co-expression clustering
PPI network computation
Sequence motif searching
GO enrichment computation
Automatic GO annotation
Miscellaneous
Enzyme kinetics computation
Computation of isothermal tiling array probes
iTRAQ data processing
Protein structure and interaction prediction
開源專案
我們維護以下三個開源專案:
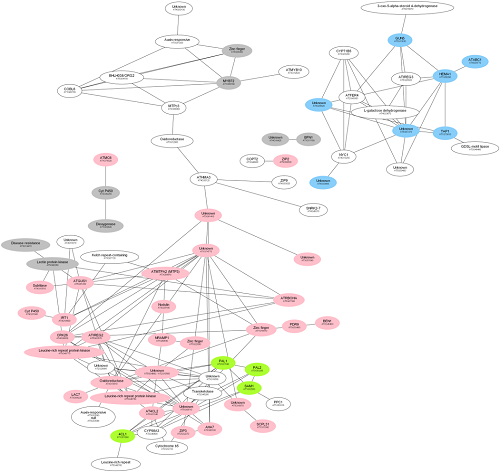
1. MACCU (Multi-Array Correlation Computation Utility, https://github.com/wdlingit/maccu ):計算並比較共表現網路(co-expression network),進一步推論組織專一之基因模組。
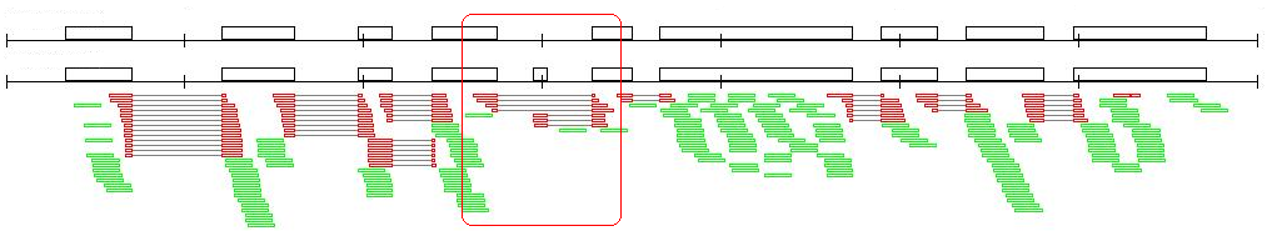
2. RackJ (Read Analysis & Comparison Kit in Java, http://rackj.sourceforge.net/ ):根據RNAseq資料,計算基因表現量,並推論與樣本有關之alternative-splicing事件。
3. GOBU (Gene Ontology Browsing Utility, https://gobu.sourceforge.io/ ):提供友善的圖形化使用者介面以操作基因本體論資料,包括快速計算富集分析。
Selected Publications
Huang CK, Lin WD, Wu SH (2022) An improved repertoire of splicing variants and their potential roles in Arabidopsis photomorphogenic development. Genome Biol. 9;23(1):50.
Hsieh EJ, Lin WD, Schmidt W (2022) Genomically Hardwired Regulation of Gene Activity Orchestrates Cellular Iron Homeostasis in Arabidopsis. RNA Biol. 19(1):143-161.
Kanno T, Venhuizen P, Wen TN, Lin WD, Chiou P, Kalyna M, Matzke AJM, Matzke M (2018) PRP4KA, a Putative Spliceosomal Protein Kinase, Is Important for Alternative Splicing and Development in Arabidopsis thaliana. Genetics. 210(4):1267-1285.
Salazar-Henao JE, Lin WD, Schmidt W (2016) Discriminative gene co-expression network analysis uncovers novel modules involved in the formation of phosphate deficiency-induced root hairs in Arabidopsis. Sci Rep 6:26820.
Kanno T, Lin WD, Fu JL, Wu MT, Yang HW, Lin SS, Matzke AJ, Matzke M (2016 Identification of Coilin Mutants in a Screen for Enhanced Expression of an Alternatively Spliced GFP Reporter Gene in Arabidopsis thaliana. Genetics 203(4):1709-20.
Sasaki T, Kanno T, Liang SC, Chen PY, Liao WW, Lin WD, Matzke AJ, Matzke M (2015) An Rtf2 domain-containing protein influences pre-mRNA splicing and is
essential for embryonic development in Arabidopsis thaliana. Genetics 200(2):523-35.
Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, López-Millán AF, and Schmidt W. (2013) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by arabidopsis and Medicago truncatula. Plant Physiology. 162:1473-85.
Li W, Lin WD, Ray P, Lan P, and Schmidt W. (2013) Genome-wide detection of condition-sensitive alternative splicing in Arabidopsis roots. Plant Physiology. 162:1750-63.
Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, and Wu SH.. (2013) Translational landscape of photomorphogenic Arabidopsis. Plant Cell. 25:3699-710.
Lan P, Li WF, Lin WD, Santi S, and Schmidt W. (2013) Mapping gene activity of Arabidopsis root hairs. Genome Biology. Genome Biology. 14:R67.
Chen YT, Shen CH, Lin WD, Chu HA, Huang BL, Kuo CI, Yeh KW, Huang LC, and Chang IF. (2013) Small RNAs of Sequoia sempervirens during rejuvenation and phase change. Plant Biology. 15:27-36.
Sharma S, Lin WD, Villamor JG, and Verslues PE. (2013) Divergent low water potential response in Arabidopsis thaliana accessions Landsberg erecta and Shahdara. Plant Cell & Environment 36: 994-1008.
Chen YC, Chen YC, Lin WD, Hsiao CD, Chiu HW, and Ho JM. (2012) Bio301: A Web-based EST annotation pipeline that facilitates functional comparison studies. ISRN Bioinformatics. (doi:10.5402/2012/139842)
Lin WD, Liao YY, Yang TJ, Pan CY, Buckhout TJ, and Schmidt W. (2011) Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiology. 155:1383-402.