
Tu, Shih-Long (涂世隆)
Research Fellow
- Ph.D., Life Science, National Defense Medical Center, Taiwan
- Molecular mechanism of light sensing and pre-mRNA splicing in plants
- tsl@gate.sinica.edu.tw
- tsl@as.edu.tw
- +886-2-2787-1045 (Lab: A321)
- +886-2-2787-1167 (Office: A342)
- Academia Sinica Archive
- ORCID
- Web of Science (WOS)
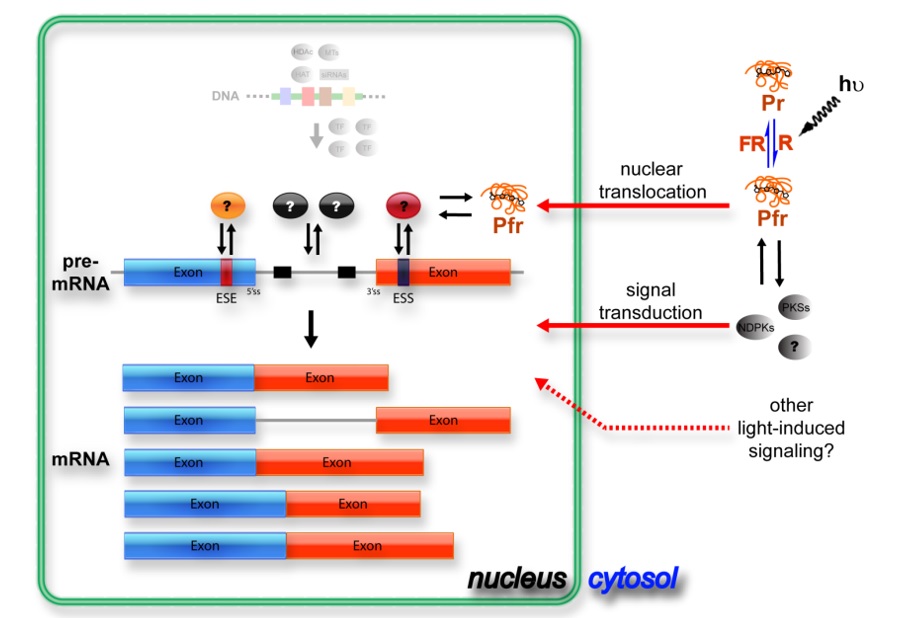
Plant growth and development are precisely and efficiently modulated in response to environmental factors. Regulatory processes for responding to environmental changes mainly involve signal perception, transduction, metabolic responses and downstream gene regulation. Interactions between environmental signals and gene regulation at chromatin, transcription, translation and post-translation levels have been well studied in recent years. However, information about regulation at the precursor mRNA (pre-mRNA) splicing step is relatively lacking.
Alternative splicing (AS) is a widespread mechanism in eukaryotes that generate 2 or more mRNAs from the same pre-mRNA by using different splice sites. Splice site selection is influenced by regulatory cis-acting sequences and trans-acting factors. Regulatory cis elements are recognized by splicing regulators to recruit spliceosomal components and initiate splicing reactions. Although AS tremendously increases transcriptome complexity and proteome diversity in the cell, whether it contributes to biological processes in plants, such as stress responses, is still little elucidated.
My laboratory has been taking systems biology approaches for studying plant gene expression in responding to environmental changes. In the case of light, we found a large number of AS events can be differentially regulated. Photoreceptors are involved in the regulation of light-responsive AS. We are investigating the molecular mechanism of light-regulated AS in plants by taking biochemistry and systems biology approaches. The final goal will be applying our understanding to modulate growth and development of agriculturally important crops and identify novel traits through regulating pre-mRNA splicing.
- Chang, P., Hsieh, H.-Y., and Tu, S.-L. (2022). A novel U1 snRNP component RBP45d regulates temperature-responsive flowering in Arabidopsis thaliana. Plant Cell. 34, 834–851.
- Cheng, Y.-L., Hsieh, H.-Y., and Tu, S.-L. (2021). A new method to identify global targets of RNA-binding proteins in plants. bioRxiv. doi: 10.1101/2021.06.11.448000.
- Wang, C.-C., Hsieh, H.-Y., Hsieh, H-L. and Tu, S.-L. (2021). The chromatin adaptor PpMRG1 interacts with H3K36me3 and regulates light-responsive alternative splicing. Plant Physiol. 185, 1229-1241.
- Petrillo, E., Kalyna, M., Mandadi, K. K., Tu, S.-L. and Simpson, C. G. (2020) Editorial: Alternative splicing regulation in plants. Front. Plant Sci.11, 913.
- Shih, C.-J., Chen, H.-W., Hsieh, H.-Y., Chiu, F.-Y., Chen, Y.-R. and Tu, S.-L. (2019). Heterogeneous nuclear ribonucleoprotein H1 coordinates with phytochrome and the U1 snRNP complex to regulate alternative splicing in Physcomitrella patens. Plant Cell. 31, 2510-2524.
- Lin, B.-Y., Shih, C.-J., and Tu, S.-L. (2019). Phytochrome coordinates with a hnRNP to regulate alternative splicing via an exonic splicing silencer. Plant Physiol. 182, 243-254.
- Chang, Y.-M., Lin, H.-H., Liu, W.-Y., Yu, C.-P., Chen, H.-J., Wartini P. P., Kao, Y.-Y., Wu, Y.-H., Lin, J.-J., Lu, M.-Y., Tu, S.-L., Wu, S.-H., Shiu, S.-H., Ku, M.-S., and Li, W.-H. (2019). Comparative transcriptomics method to infer gene coexpression networks and its applications to maize and rice leaf transcriptomes. Proc. Natl. Acad. Sci. USA. 116, 3091-3099.
- Cheng, Y.-L. and Tu, S.-L. (2018). New insights of light-regulated alternative splicing. Plant. Cell. Physiol. Invited Review. 59, 1104-1110.
- C.-F., Yu, C.-P., Wu, Y.-H., Lu, M.-Y., Tu, S.-L., Wu, S.-H., Shiu, S.-H, Ku, M.-S., and Li, W.-H. (2017). Elevated auxin biosynthesis and transport underlie high vein density in C4 leaves. Proc. Natl. Acad. Sci. USA. 114, E6884-E6891.
- Huang, C.-F., Chang, Y.-M., Lin, J.-J., Yu, C.-P., Lin, H.-H., Liu, W.-Y., Yeh, S., Tu, S.-L., Wu, S.-H., Ku, M.-S.*, and Li, W.-H.* (2016). Insights into the regulation of C4 leaf development from comparative transcriptomic analysis. Curr. Opin. Plant Biol. 30, 1-10.
- Chang, C.-Y., Lin, W.-D., and Tu, S.-L. (2014). Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 165, 826-840.
- Wu, H.-P., Su, Y.-s., Chen, H.-C., Chen, Y.-R., Wu, C.-C., Lin, W.-D., and Tu, S.-L. (2014). Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 15, R10.
- Chen, Y.-R., Su, Y.-s., and Tu, S.-L. (2012). Distinct phytochrome actions in non-vascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proc. Natl. Acad. Sci. USA. 109, 8310-8315.
- Chiu, F.-Y., Chen, Y.-R., and Tu, S.-L. (2010). Electrostatic interaction of phytochromobilin synthase and ferredoxin for biosynthesis of phytochrome chromophore. J. Biol. Chem. 285, 5056-5065.
- Tu, S.-L., Chen, H.-C., Ku, L.-W. (2008). Mechanistic studies of the phytochromobilin synthase HY2 from Arabidopsis. J. Biol. Chem. 283, 27555-27564.


You-Liang Cheng Postdoctoral Fellow yeoliang@gate.sinica.edu.tw

Hsin-Yu Hsieh Research Assistant hsinyuh@gate.sinica.edu.tw

Ping Chang Graduate Student greenfavor@gate.sinica.edu.tw

Bou-Yun Lin Graduate Student rogerbyl0704@gate.sinica.edu.tw

Chueh-Ju Shih Graduate Student positivegloria@gate.sinica.edu.tw

Jainn-Zang Wang Graduate Student taiwanod@gate.sinica.edu.tw
International
- 2017: AOSP Award for Young Scientists, Asia and Oceania Society for Photobiology (AOSP)
Domestic
- 2022: Outstanding Research Award, National Science and Technology Council
- 2013: Junior Research Investigator Award, Academia Sinica
- 2013: Outstanding Scholars Program , National Science Council
- 2012: The Shang-Fa Yang Young Scientist Award, The Shang-Fa Yang Memorial Foundation
- 2010: Career Development Award, Academia Sinica