
郭志鴻 (Kuo, Chih-Horng)
學術副所長
研究員
- 美國喬治亞大學遺傳學系 博士
- 共生細菌的演化與功能性基因體學
- chk@gate.sinica.edu.tw
- chk@as.edu.tw
- +886-2-2787-1044 (Lab: R224)
- +886-2-2787-1127 (Office: R224)
- Lab Website
- Academia Sinica Archive
- ORCID
- Web of Science (WOS)
- Google Scholar
研究主題:共生細菌的演化與功能性基因體學
在微生物的演化上,一旦一個物種發展出對真核宿主的依存性,其基因體的結構經常會有大規模的變化。為了解與這些變化相關的演化過程和其基因內容之改變,本研究室利用基因體學及轉錄體學等工具進行不同細菌物種間的比較分析。近年之研究計畫包含兩個主要的生物系統:
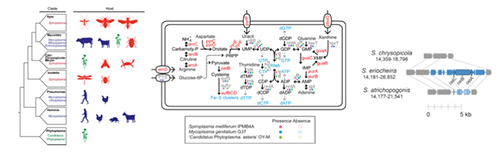
(1) 柔膜細菌綱 (Mollicutes)
柔膜細菌綱為一群不具有細胞壁之宿主依存性細菌,包括植物菌質體 (‘Candidatus Phytoplasma’; 為一群農業上重要之植物病原)、黴漿菌(Mycoplasma; 脊椎動物病原)、及螺旋菌質體 (Spiroplasma; 大多為昆蟲共生菌,少數物種為植物病原) 等代表性物種。其中螺旋菌質體屬內物種可能與宿主進行片利共生、互利共生、或依賴程度不等的寄生,因此跨物種的比較將有助了解細菌在生態棲位間的轉換及致病性的演化。針對此生物系統,本研究室的短期目標為調查可解釋物種間表現型差異之遺傳機制,而長期目標則是累積足夠的基因體資料以進行完整的跨屬分析。
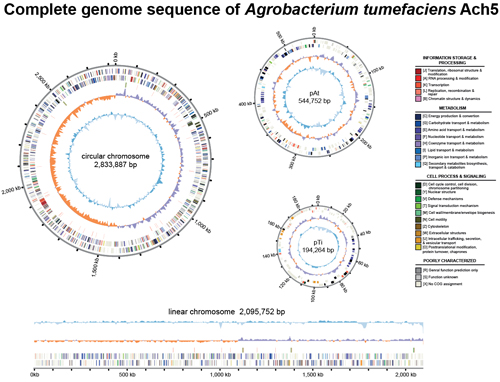
(2) 農桿菌屬 (Agrobacterium)
農桿菌屬為一群與植物相關的細菌,由於這群細菌在植物基因轉殖上的應用和其在田間對農作物所產生的危害,在生物科技和農業的發展上都具有相當的重要性。本研究室與所內賴爾珉老師合作,共同進行農桿菌屬內物種的研究,藉以探討此屬內不同小群菌株間的遺傳歧異度、基因體演化、及功能性分析。
Reviews
- Weisberg AJ*, Wu Y, Chang JH, Lai EM, Kuo CH* (2023) Virulence and ecology of agrobacteria in the context of evolutionary genomics. Annual Review of Phytopathology 61:1-23. DOI: 10.1146/annurev-phyto-021622-125009
- Lo WS, Huang YY, Kuo CH* (2016) Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiology Reviews 40:855-874. DOI: 10.1093/femsre/fuw028
Research articles
- Nishino T#, Moriyama M#*, Mukai H#, Tanahashi M#, Hosokawa T, Chang HY, Tachikawa S, Nikoh N, Koga R, Kuo CH, Fukatsu T* (2025) Defensive fungal symbiosis on insect hindlegs. Science 390:279-283. DOI: 1126/science.adp6699 #Equal contribution [News by Erik Stokstad on Science DOI: 10.1126/science.zlhmcem] [Featured on Academia Sinica website]
- Chiu YC, Lin YC, Pao SH, Chen YK, Liao PQ, Mejia HM, Sheue CR, Kuo CH*, Yang JY* (2025) Comprehensive genomic and functional characterization of a phytoplasma associated with root retardation, early bolting, witches’-broom, and phyllody in daikon (Raphanus sativus). Frontiers in Microbiology 16:1654928. DOI: 10.3389/fmicb.2025.1654928
- Wu Y, Chang HY, Wu CH, Lai EM*, Kuo CH* (2025) Comparative transcriptomics reveals context- and strain-specific regulatory programs of Agrobacterium during plant colonization. Microbial Genomics 11:001485. DOI: 10.1099/mgen.0.001485 [bioRxiv preprint DOI: 10.1101/2025.05.27.656240]
- Chen CNN#, Lin KM#, Lin YC#, Chang HY, Yong TC, Chiu YF, Kuo CH*, Chu HA* (2025) Comparative genomic analysis of a novel heat-tolerant and euryhaline strain of unicellular marine cyanobacterium Cyanobacterium DS4 from a high-temperature lagoon. BMC Microbiology 25:279. DOI: 10.1186/s12866-025-03993-7 [bioRxiv preprint DOI: 10.1101/2025.01.17.633688] #Equal contribution
- Seruga Music M*, Polak B, Drcelic M, Pei SC, Kuo CH* (2025) Sequencing and comparative analyses of ‘Candidatus Phytoplasma solani’ genomes reveal diversity of effectors and potential mobile units. Microbial Genomics 11:001401. DOI: 10.1099/mgen.0.001401
- Lopez-Agudelo JC#, Goh FJ#, Tchabashvili S, Huang YS, Huang CY, Lee KT, Wang YC, Wu Y, Chang HX, Kuo CH, Lai EM, Wu CH* (2025) Rhizobium rhizogenes A4-derived strains mediate hyper-efficient transient gene expression in Nicotiana benthamiana and other solanaceous plants. Plant Biotechnology Journal published online. DOI: 10.1111/pbi.70083 [bioRxiv preprint DOI: 1101/2024.11.30.626145] #Equal contribution [Featured on Academia Sinica website]
- Chang HY, Yen HC, Chu HA, Kuo CH* (2024) Population genomics of a thermophilic cyanobacterium revealed divergence at subspecies level and possible adaptation genes. Botanical Studies 65:35. DOI: 10.1186/s40529-024-00442-y [bioRxiv preprint DOI: 10.1101/2024.08.22.609105]
- Yan XH, Pei SC, Yen HC, Blanchard A, Sirand-Pugnet P, Baby V, Gasparich GE, Kuo CH* (2024) Delineating bacterial genera based on gene content analysis: a case study of the Mycoplasmatales–Entomoplasmatales clade within the class Mollicutes. Microbial Genomics 10:001321. DOI: 10.1099/mgen.0.001321 [bioRxiv preprint DOI: 10.1101/2024.08.17.608393]
- Wang SC, Chen AP, Chou SJ, Kuo CH*, Lai EM* (2023) Soil inoculation and blocker-mediated sequencing show effects of the antibacterial T6SS on agrobacterial tumorigenesis and gallobiome. mBio 14:e00177-23. DOI:10.1128/mbio.00177-23 [bioRxiv preprint DOI: 10.1101/2022.10.28.514271] [Featured on Academia Sinica website]
- Cheng YI, Lin YC, Leu JY, Kuo CH*, Chu HA* (2022) Comparative analysis reveals distinctive genomic features of Taiwan hot-spring cyanobacterium Thermosynechococcus TA-1. Frontiers in Microbiology 13:932840. DOI: 10.3389/fmicb.2022.932840
- Huang CT, Cho ST, Tan CM, Chiu YC, Yang JY*, Kuo CH* (2022) Comparative genome analysis of ‘Candidatus Phytoplasma luffae’ reveals the influential roles of potential mobile units in phytoplasma evolution. Frontiers in Microbiology 13:773608. DOI: 10.3389/fmicb.2022.773608 [bioRxiv preprint DOI: 10.1101/2021.09.09.459700]
- Chou L#, Lin YC#, Haryono M, Santos MNM, Cho ST, Weisberg AJ, Wu CF, Chang JH, Lai EM, Kuo CH* (2022) Modular evolution of secretion systems and virulence plasmids in a bacterial species complex. BMC Biology 20:16. DOI: 10.1186/s12915-021-01221-y [bioRxiv preprint DOI: 10.1101/2021.05.20.444927] #Equal contribution [Featured on Academia Sinica website]
- Castillo AI, Tsai CW, Su CC, Weng LW, Lin YC, Cho ST, Almeida RPP, Kuo CH* (2021) Genetic differentiation of Xylella fastidiosa following the introduction into Taiwan. Microbial Genomics 7:000727. DOI: 10.1099/mgen.0.000727 [bioRxiv preprint DOI: 10.1101/2021.03.24.436723]
- Tan CM, Lin YC, Li JR, Chien YY, Wang CJ, Chou L, Wang CW, Chiu YC, Kuo CH*, Yang JY* (2021) Accelerating complete phytoplasma genome assembly by immunoprecipitation-based enrichment and MinION-based DNA sequencing for comparative analyses. Frontiers in Microbiology 12:766221. DOI: 10.3389/fmicb.2021.766221
- Huang W, MacLean A, Sugio A, Maqbool A, Busscher M, Cho ST, Kamoun S, Kuo CH, Immink RGH, Hogenhout SA* (2021) Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell 184:5201-5214. [Issue Cover] DOI: 10.1016/j.cell.2021.08.029 [bioRxiv preprint DOI: 10.1101/2021.02.15.430920]
- Weng LW, Lin YC, Su CC, Huang CT, Cho ST, Chen AP, Chou SJ, Tsai CW*, Kuo CH* (2021) Complete genome sequence of Xylella taiwanensis and comparative analysis of virulence gene content with Xylella fastidiosa. Frontiers in Microbiology 12:684092. DOI: 10.3389/fmicb.2021.684092 [bioRxiv preprint DOI: 10.1101/2021.03.08.434500]
- Cho ST, Kung HJ, Huang W, Hogenhout SA, Kuo CH* (2020) Species boundaries and molecular markers for classification of 16SrI phytoplasmas inferred by genome analysis. Frontiers in Microbiology 11:1531. DOI: 10.3389/fmicb.2020.01531 [bioRxiv preprint DOI: 10.1101/2020.01.31.928135]
- Weisberg AJ, Davis EW, Tabima J, Belcher MS, Miller M, Kuo CH, Loper JE, Grünwald NJ, Putnam ML, Chang JH* (2020) Unexpected conservation and global transmission of agrobacterial virulence plasmids. Science 368:eaba5256. DOI: 10.1126/science.aba5256
- Cheng YI, Chou L, Chiu YF, Hsueh HT, Kuo CH*, Chu HA* (2020) Comparative genomic analysis of a novel strain of Taiwan hot-spring cyanobacterium Thermosynechococcus CL-1. Frontiers in Microbiology 11:82. DOI: 10.3389/fmicb.2020.00082
- Cho ST, Lin CP, Kuo CH* (2019) Genomic characterization of the periwinkle leaf yellowing (PLY) phytoplasmas in Taiwan. Frontiers in Microbiology 10:2194. DOI: 10.3389/fmicb.2019.02194
- Haryono M, Cho ST, Fang MJ, Chen AP, Chou SJ, Lai EM, Kuo CH* (2019) Differentiations in gene content and expression response to virulence induction between two Agrobacterium Frontiers in Microbiology 10:1554. DOI: 10.3389/fmicb.2019.01554 [Recommendation by Vitaly Citovsky on Faculty Opinions DOI: 10.3410/f.736286615.793563521]
- Wu CF, Santos MNM, Cho ST, Chang HH, Tsai YM, Smith DA, Kuo CH*, Chang JH*, Lai EM* (2019) Plant pathogenic Agrobacterium tumefaciens strains have diverse type VI effector-immunity pairs and vary in in-planta Molecular Plant-Microbe Interactions 32:961-971. DOI: 10.1094/MPMI-01-19-0021-R [Editor's Pick]
- Seruga Music M*, Samarzija I, Hogenhout SA*, Haryono M, Cho ST, Kuo CH* (2019) The genome of ‘Candidatus Phytoplasma solani’ strain SA-1 is highly dynamic and prone to adopting foreign sequences. Systematic and Applied Microbiology 42:117-127. DOI: 10.1016/j.syapm.2018.10.008
- Haryono M, Tsai YM, Lin CT, Huang FC, Ye YC, Deng WL, Hwang HH*, Kuo CH* (2018) Presence of an Agrobacterium-type tumor-inducing plasmid in Neorhizobium NCHU2750 and the link to phytopathogenicity. Genome Biology and Evolution 10:3188-3195. DOI: 10.1093/gbe/evy249
- Lo KJ, Lin SS, Lu CW, Kuo CH*, Liu CT* (2018) Whole-genome sequencing and comparative analysis of two plant-associated strains of Rhodopseudomonas palustris (PS3 and YSC3). Scientific Reports 8:12769. DOI: 10.1038/s41598-018-31128-8
- Lo WS, Gasparich GE, Kuo CH* (2018) Convergent evolution among ruminant-pathogenic Mycoplasma involved extensive gene content changes. Genome Biology and Evolution 10:2130-2139. DOI: 10.1093/gbe/evy172
- Tsai YM, Chang A, Kuo CH* (2018) Horizontal gene acquisitions contributed to genome expansion in insect-symbiotic Spiroplasma clarkii. Genome Biology and Evolution 10:1526-1532. DOI: 10.1093/gbe/evy157
- Lo WS, Kuo CH* (2017) Horizontal acquisition and transcriptional integration of novel genes in mosquito-associated Spiroplasma. Genome Biology and Evolution 9:3246-3259. DOI: 10.1093/gbe/evx244
- Orlovskis Z, Canale MC, Haryono M, Lopes JRS, Kuo CH*, Hogenhout S* (2017) A few sequence polymorphisms among isolates of maize bushy stunt phytoplasma associate with organ proliferation symptoms in infected maize plants. Annals of Botany 119:869-884. DOI: 10.1093/aob/mcw213
- Bondage DD, Lin JS, Ma LS, Kuo CH, Lai EM* (2016) VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor–effector complex. Proceedings of the National Academy of Sciences of the United States of America 113:E3931-E3940. DOI: 10.1073/pnas.1600428113 [Recommendation by Alain Filloux on Faculty Opinions DOI: 10.3410/f.726430927.793522872]
- Cho ST, Chang HH, Egamberdieva D, Kamilova F, Lugtenberg B, Kuo CH* (2015) Genome analysis of Pseudomonas fluorescens PCL1751: a rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLOS ONE 10:e0140231. DOI: 10.1371/journal.pone.0140231
- Lo WS, Gasparich GE, Kuo CH* (2015) Found and lost: the fates of horizontally acquired genes in arthropod-symbiotic Spiroplasma. Genome Biology and Evolution 7:2458-2472. DOI: 10.1093/gbe/evv160
- McGaugh SE*, Bronikowski AM*, Kuo CH*, Reding DM, Addis EA, Flagel LE, Janzen FJ, Schwartz TS* (2015) Rapid molecular evolution across amniotes of the IIS/TOR network. Proceedings of the National Academy of Sciences of the United States of America 112:7055-7060. DOI: 10.1073/pnas.1419659112
- Ku C, Lo WS, Kuo CH* (2014) Molecular evolution of the actin-like MreB protein gene family in wall-less bacteria. Biochemical and Biophysical Research Communications 446:927-932. DOI: 10.1016/j.bbrc.2014.03.039
- Chang TH, Lo WS, Ku C, Chen LL, Kuo CH* (2014) Molecular evolution of the substrate utilization strategies and putative virulence factors in mosquito-associated Spiroplasma Genome Biology and Evolution 6:500-509. DOI: 10.1093/gbe/evu033
- Lo WS, Ku C, Chen LL, Chang TH, Kuo CH* (2013) Comparison of metabolic capacities and inference of gene content evolution in mosquito-associated Spiroplasma diminutum and taiwanense. Genome Biology and Evolution 5:1512-1523. DOI: 10.1093/gbe/evt108
- Ku C, Lo WS, Chen LL, Kuo CH* (2013) Complete genomes of two dipteran-associated spiroplasmas provided insights into the origin, dynamics, and impacts of viral invasion in Spiroplasma. Genome Biology and Evolution 5:1151-1164. DOI: 10.1093/gbe/evt084
- Lo WS, Chen LL, Chung WC, Gasparich GE, Kuo CH* (2013) Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genomics 14:22. DOI: 10.1186/1471-2164-14-22
- Chung WC, Chen LL, Lo WS, Lin CP, Kuo CH* (2013) Comparative analysis of the peanut witches’-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLOS ONE 8:e62770. DOI: 10.1371/journal.pone.0062770
- Chen LL, Chung WC, Lin CP, Kuo CH* (2012) Comparative analysis of gene content evolution in phytoplasmas and mycoplasmas. PLOS ONE 7:e34407. DOI: 10.1371/journal.pone.0034407
國際
- 2023 Derrick Edward Award - 國際黴漿菌學學會(IOM)
- 2021 羅伯特.惠特康獎 - 國際黴漿菌學會
國內
- 2020 優秀年輕學者研究計畫 - 科技部
- 2019 林秋榮植物科學創新研究獎 - 財團法人林秋榮植物科學教育基金會
- 2017 年輕學者研究著作獎 - 中央研究院
- 2013 新秀獎 - 臺灣植物學會