
邢禹依 (Hsing, Yue-Ie)
特聘研究員
- 美國伊利諾大學博士
- 水稻基因組研究與大豆種子成熟蛋白研究
- bohsing@gate.sinica.edu.tw
- bohsing@as.edu.tw
- Academia Sinica Archive
研究主題:
• 製備T-DNA標記水稻突變族群及網頁
• 我們製備了以T-DNA做為標記的水稻突變族 群 (TRIM) ,以進行功能基因體研究。我負責生物資訊的部份。
• 針對TRIM突變族群進行研究
• 我們以數個此剔除及活化突變族群,進行深入研究。
• 以新世代定序方式定位與定性重要水稻突變株
• 我們針對數個水稻種子發育及幼苗生長的突變株、進行定位與定性分析。
• 以新世代定序方式研究水稻之現代品種與早期地方品系
• 我們以台灣現代品種與早期地方品系為材料,探討導入、作物化、及進一步的利用。
• 定序分析一年生野生水稻Oryza nivara
• 我們參與國際組織,目的在定序分析數個一年生野生水稻。本實驗室負責Oryza nivara 的工作。
• 解讀水稻與大豆基因組之種子成熟蛋白基因
• 種子成熟蛋白存在於許多物種(包括植物、動物及微生物),指出了它們的重要性。我們解讀水稻及大豆兩種模式植物基因體的種子成熟蛋白基因。
本實驗室負責之網頁
水稻第五條染色體定序分析 http://genome.sinica.edu.tw/index.htm
T-DNA標記水稻突變族群 http://trim.sinica.edu.tw/
種子成熟蛋白資料庫 http://ipmb.sinica.edu.tw/soja/LEA/review_list/
水稻花葯轉譯體資料 http://rice.sinica.edu.tw/rice_anther/
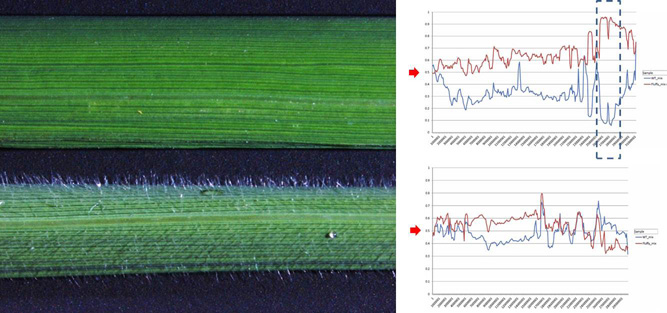
- Wei FJ, Kuang LY, Oung HM, Cheng SY, Wu HP, Huang LT, Tseng YT, Chiou WY, Hsieh-Feng V, Chung CH, Yu SM, Lee LY, Gelvin SB, Hsing YIC*. 2016. Somaclonal variation does not preclude using rice transformants for genetic screening. Plant Jour. (in press)
- Lo SF, Fan MJ, Hsing YI, Chen LJ, Chen S, Wen IC, Liu YL, Chen KT, Jing MR, Lin MK, Rao MY, Yu LC, Ho THD, Yu SM*. 2016. Genetic resources offer efficient tools for rice functional genomics research. Plant, Cell & Environment (in press)
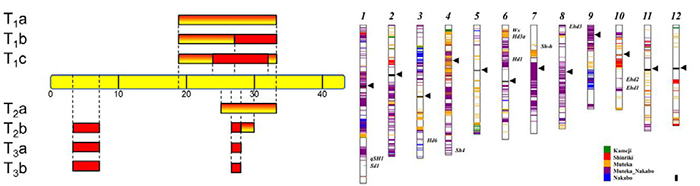
- Wei FJ, Tsai YC, Wu HP, Huang LT, Chen YC, Chen YF, Wu CC, Tseng YT, Hsing YIC*. 2016. Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 242: 187-194
- Copetti D, Zhang J, Baidouri ME, Gao D, Wang J, Barghini E, Cossu RM, Angelova A, Maldonado CE, Roffler S, Ohyanagi H, Wicker T, Fan G, Zuccolo A, Chen M, Oliveira AC, Han B, Henry R, Hsing YI, Kurata N, Wang W, Jackson SA, Panaud O, Wing RA*. 2015. Oryza Repeat Database: a Resource for Genus-wide Rice Genomics and Evolutionary Biology. BMS Genomics 16:538
- Baldrich P, Campo S, Wu MT, Hsing YIC, San Segundo B*. 2015. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biology 12: 847--863.
- Dixit S, Biswal AK, Min A, Henry A, Oane RH, Raorane ML, Longkumer T, Pabuayon IM, Mutte SK, Varadarajan AR, Miro B, Enriquez BA, Serafin L, Lozada M, Tsai YC, Sundaravelpandian K, Crowell S, Slamet-Loedin I, Hsing YIC, McCouch SR, Kumar A, Kohli A*. 2015. A multi-trait multi-gene region underpins a QTL for rice yield under drought. Scientific Report 5:15183.
- Chen H, Chung MC, Tsai YC, Wei FJ, Wu CC, Hsieh, JS, YIC Hsing*. 2015. Distribution of new satellites and simple sequence repeats in annual and perennial Glycine species. Botanical Studies 56:22
- Hour ZY, Wu MT, Lu SH, Hsing YIC, Chen HM*. 2014. Beyond the cleavage products of small regulatory RNAs: unraveling the complexity of plant RNA degradome data. BMC Genomics. 15:15.
- Hsu YC, Tseng MC, Wu YP, Lin MY, Wei FJ, Hwu KK, Hsing YI, Lin YR*. 2014. Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Molecular Breeding. 34:655–673
- Wei FJ, Droc G, Guiderdoni E, Hsing YIC*. 2013. International Consortium of Rice Mutagenesis: resources and beyond. Rice 6:39
- Shih, MD, TY Hsieh, WT Jian, MT Wu, SJ Yang, FA Hoekstra, YIC Hsing.* 2012. Functional studies of soybean (Glycine max L.) seed LEA proteins, GmPM6, GmPM11, and GmPM30, by CD and FTIR spectroscopy. Plant Sci. 196: 152-159.
- Chern CG, MJ Fan, SC Huang, SM Yu, FJ Wei, CC Wu, A Trisiriroj, MH Lai, YIC Hsing*. 2010. Methods for rice phenomics studies. In: Plant Reverse Genetics: Methods and Protocols, Methods in Molecular Biology, vol. 678, A Pereira (ed.) Humana Press. pp.129-138.
- Shih MD, FA Hoekstra, YIC Hsing*. 2008. Late embryogenesis abundant proteins. Adv. Bot. Res. 48: 211-255.
國際
- 2022 終身成就獎 - 國際水稻功能基因組會議
- 2009 美國科學促進學會會士 - 美國科學促進學會
- 2008 花喇子模國際科學獎 - 伊朗科學暨科技研究組織
- 2007 臺法科技獎 - 法國自然科學院與科技部
國內
- 2022 終身成就獎 - 臺灣植物學會
- 2012 優秀農業人員獎 - 行政院農業委員會
- 2011 深耕計畫 - 中央研究院