
Hsing, Yue-Ie (邢禹依)
Distinguished Research Fellow
- Ph. D., Dept. AgronomyUniversity of Illinois, USA
- Rice genomics study and research on soybean seed maturation proteins
- bohsing@gate.sinica.edu.tw
- bohsing@as.edu.tw
- Academia Sinica Archive
Research areas
• Construction a Tagged Rice Population, TRIM, for functional genomics analysis We develop an indexed mutant population for rice functional
genomics studies and I'm responsible for the bioinformatics part.
• Utilization/studies of the TRIM mutant resources With the huge collection of activated and/or knocked-out mutants and detailed phenomics data,
my lab picks up some mutants and performs detail analysis.
• Next generation sequencing (NGS) mapping and characterization of important rice mutants We work on the gene finding/characterization of several
rice mutants related to seed development and seedling growth.
• NGS analysis of rice modern varieties and land races in Taiwan We work on the modern varieties, the recent and very old land races in Taiwan,
with emphasis on introgression,domestication, and utilization.
• Sequencing of Oryza nivara, an annual wild rice We joined the international consortium with the goal to decode several wild rice species.
My lab is responsible for an annual AA genome, Oryza nivara .
• Re-annotation of genes encoding LEA proteins in the rice and soybean genomes The extensive distribution of LEA genes among diverse taxa impliesthat
these genes might be primitive yet important and therefore maintained by these species. We work on the re-annotation of LEA genes in two important
model plants.
Websites related to my research
• Rice chromosome 5 sequencing project
http://genome.sinica.edu.tw/index.htm
• A resource for rice functional genomics study
http://trim.sinica.edu.tw/
• Late embryogenesis abundant proteins
http://ipmb.sinica.edu.tw/soja/LEA/review_list/
• Transcriptome analysis of rice anthers
http://rice.sinica.edu.tw/rice_anther/
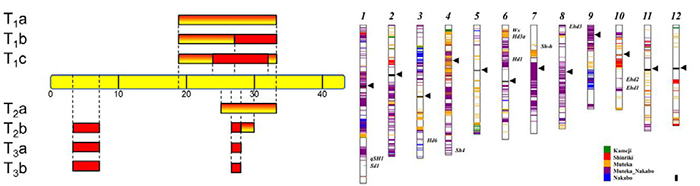
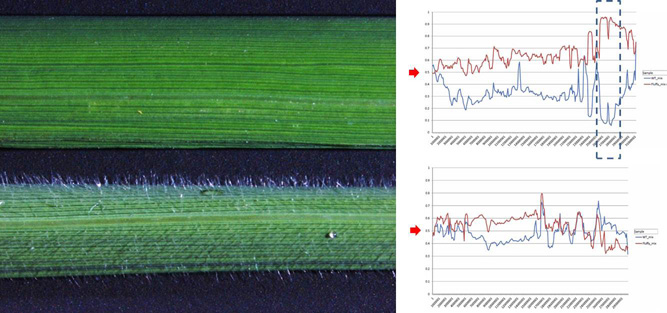
- Wei FJ, Kuang LY, Oung HM, Cheng SY, Wu HP, Huang LT, Tseng YT, Chiou WY, Hsieh-Feng V, Chung CH, Yu SM, Lee LY, Gelvin SB, Hsing YIC*. 2016. Somaclonal variation does not preclude using rice transformants for genetic screening. Plant Jour. (in press)
- Lo SF, Fan MJ, Hsing YI, Chen LJ, Chen S, Wen IC, Liu YL, Chen KT, Jing MR, Lin MK, Rao MY, Yu LC, Ho THD, Yu SM*. 2016. Genetic resources offer efficient tools for rice functional genomics research. Plant, Cell & Environment (in press)
- Wei FJ, Tsai YC, Wu HP, Huang LT, Chen YC, Chen YF, Wu CC, Tseng YT, Hsing YIC*. 2016. Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 242: 187-194
- Copetti D, Zhang J, Baidouri ME, Gao D, Wang J, Barghini E, Cossu RM, Angelova A, Maldonado CE, Roffler S, Ohyanagi H, Wicker T, Fan G, Zuccolo A, Chen M, Oliveira AC, Han B, Henry R, Hsing YI, Kurata N, Wang W, Jackson SA, Panaud O, Wing RA*. 2015. Oryza Repeat Database: a Resource for Genus-wide Rice Genomics and Evolutionary Biology. BMS Genomics 16:538
- Baldrich P, Campo S, Wu MT, Hsing YIC, San Segundo B*. 2015. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biology 12: 847--863.
- Dixit S, Biswal AK, Min A, Henry A, Oane RH, Raorane ML, Longkumer T, Pabuayon IM, Mutte SK, Varadarajan AR, Miro B, Enriquez BA, Serafin L, Lozada M, Tsai YC, Sundaravelpandian K, Crowell S, Slamet-Loedin I, Hsing YIC, McCouch SR, Kumar A, Kohli A*. 2015. A multi-trait multi-gene region underpins a QTL for rice yield under drought. Scientific Report 5:15183.
- Chen H, Chung MC, Tsai YC, Wei FJ, Wu CC, Hsieh, JS, YIC Hsing*. 2015. Distribution of new satellites and simple sequence repeats in annual and perennial Glycine species. Botanical Studies 56:22
- Hour ZY, Wu MT, Lu SH, Hsing YIC, Chen HM*. 2014. Beyond the cleavage products of small regulatory RNAs: unraveling the complexity of plant RNA degradome data. BMC Genomics. 15:15.
- Hsu YC, Tseng MC, Wu YP, Lin MY, Wei FJ, Hwu KK, Hsing YI, Lin YR*. 2014. Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Molecular Breeding. 34:655–673
- Wei FJ, Droc G, Guiderdoni E, Hsing YIC*. 2013. International Consortium of Rice Mutagenesis: resources and beyond. Rice 6:39
- Shih, MD, TY Hsieh, WT Jian, MT Wu, SJ Yang, FA Hoekstra, YIC Hsing.* 2012. Functional studies of soybean (Glycine max L.) seed LEA proteins, GmPM6, GmPM11, and GmPM30, by CD and FTIR spectroscopy. Plant Sci. 196: 152-159.
- Chern CG, MJ Fan, SC Huang, SM Yu, FJ Wei, CC Wu, A Trisiriroj, MH Lai, YIC Hsing*. 2010. Methods for rice phenomics studies. In: Plant Reverse Genetics: Methods and Protocols, Methods in Molecular Biology, vol. 678, A Pereira (ed.) Humana Press. pp.129-138.
- Shih MD, FA Hoekstra, YIC Hsing*. 2008. Late embryogenesis abundant proteins. Adv. Bot. Res. 48: 211-255.
International
- 2022: Pioneer Award for Outstanding and Sustained Contributions to the Advancement of Rice Genome Biology, International Symposium on Rice Functional Genomics (ISRFG)
- 2009: Fellow of AAAS, American Association for the Advancement of Science (AAAS)
- 2008: Khwarizmi International Award, Iranian Research Organization for Science and Technology
- 2007: Franco-Taiwanese Scientific Grand Prize, l’Académie des Sciences Française (FR) & Ministry of Science and Technology (TW)
Domestic
- 2022: Lifetime Achievement Award, Taiwan Society of Plant Biologists
- 2012: Distinguished Agriculturist Award, Council of Agriculture
- 2011: Investigator Award, Academia Sinica