Dr. Wan-Sheng Sunny Lo (羅椀升)
- Ph.D., Department of Biological Sciences, St. John's University, USA
Research Interests:
The main interest of my research team is to study the regulatory mechanism in chromatin modifications leading to formation of transcriptionally active or repressed complexes that modulate gene expression in various cellular activities. We have utilized budding yeast Saccharomyces cerevisiae and Arabidopsis thaliana as the experimental organisms to investigate critical issues in chromatin dynamics, transcriptional regulation and genome stability. Two research foci are:
I. Mechanistic study of histone modifications in controlling chromatin dynamics, gene expression and genome stability in response to environmental stresses
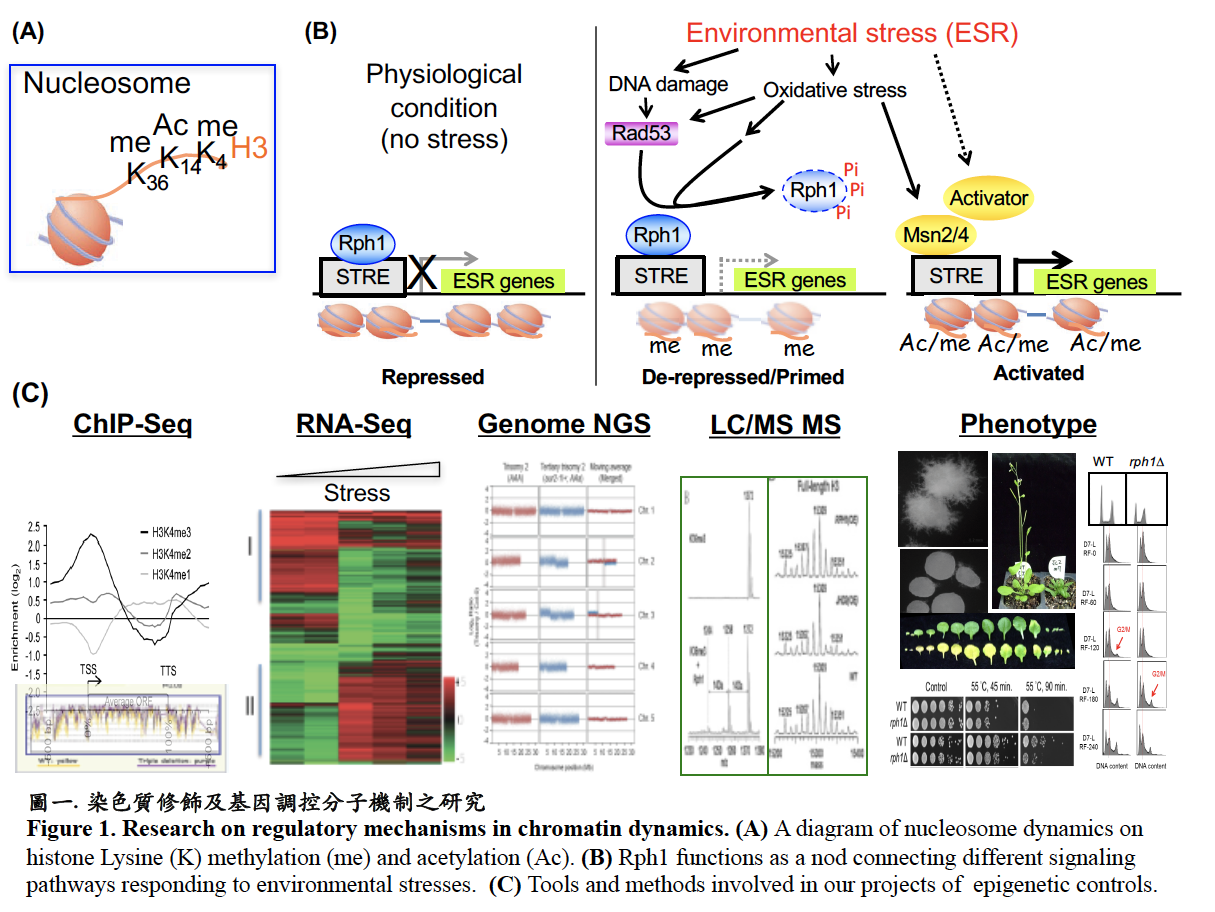
The dynamic change in chromatin structure that promotes switching between repression and activation of gene expression plays a key role in almost all aspects of cellular function. My research team was one of the first groups to identify JmjC domain-containing histone lysine demethylase (JHDM) family in the budding yeast. My current research focuses on functional characterization of certain members in the JHDM family in yeast and Arabidopsis. By using ChIP-Seq, RNA-Seq, NGS, LC/MS MS and phenotypical analysis, we investigated the interplay of various chromatin modifications involving histone H3K36 demethylase (ScRph1) and H3K4 demethylase (ScJhd2 and AtJP1) in transcriptional regulation responding to developmental signals and environmental stresses. We also revealed a role of epigenetic control of ncRNA in pseudohyphae development in budding yeast (Fig. 1).
II. Use of budding yeast to study epigenetic controls in pseudohyphae development and identify antifungal compounds by a chemical genetics strategy
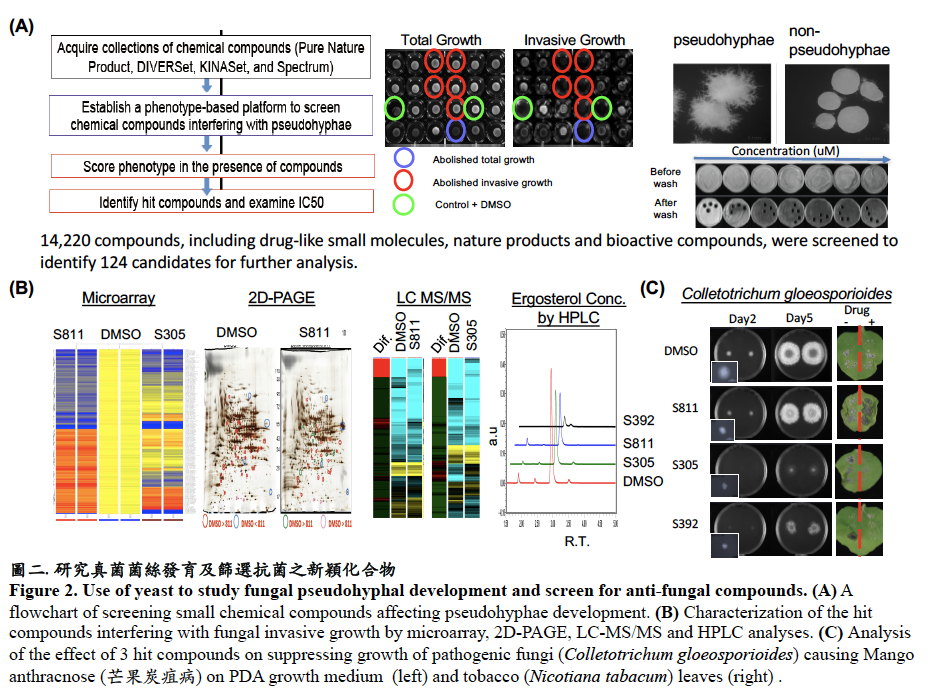
Studies of fungal virulence factors provide an in-depth understanding for systemic fungal infection and the potential to identify targets for effective anti-fungal treatments. We have used a phenotype-based chemical screening to identify small molecule compounds interfering with pseudohyphae and hyphae development and determine the mechanism of action. Results from our work are expected to provide practical scientific values to basic and medical research and offer potential applications for anti-fungal therapy (Fig. 2).
- Wang LC, Montalvo-Munoz F, Tsai YC, Liang CY, Chang CC, Lo WS*. (2015) The histone acetyltransferase, Gcn5, regulates ncRNA-ICR1 and FLO11expression for pseudohyphal development in yeast Saccharomyces cerevisiae. BioMed Research International doi:10.1155/2015/284692
- Catinot J, Huang JB, Huang PY, Tseng MY, Chen YL, Gu SY, Lo WS, Wang LC, Chen YR, Zimmerli L*. (2015) ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate - and ethylene-responsive defence genes. Plant, Cell and Environment doi:10.1111/pce.12583
- Lo KL, Wang LC, Chen IJ, Liu YC, Chung MC and Lo WS* (2014) Transcriptional consequence and impaired gametogenesis with high-grade aneuploidy in Arabidopsis thaliana. PLoS One, doi:10.1371/journal.pone.0114617.
- Liang CY, Wang LC, Lo WS * (2013) Dissociation of the H3K36 demethylase Rph1 from chromatin mediates derepression of environmental stress-response genes under genotoxic stress in Saccharomyces cerevisiae. Molecular Biology of the Cell 24(20):3251-3262.
- Chen IJ, Lo WS, Chuang JY, Cheuh CM, Fan YS, Lin YC, Wu SJ, Wang LC* (2013) Chemical genetics reveals a role of brassinolides and cellulose synthase in the hypocotyl development of etiolated Arabidopsis seedlings. Plant Science 209: 46-57.
- Liang CY, Hsu PH, Chou DF, Pan CY, Wang LC, Huang WC, Tsai MD, Lo WS * (2011) The histone H3K36 demethylase Rph1/KDM4 regulates the expression of the photoreactivation gene PHR1. Nucleic Acids Research. 39(10): 4151-4165.
- Tu S, E. Bulloch MM, Yang L, Ren C, Huang WC, Hsu PH., CH Chen CH, Liao CL, Yu HM, Lo WS *, Freitas MA*, Tsai MD* (2007) Identification of histone demethylases in Sacchromyces cerevisiae. J. Biol. Chem. 282(19): 14262-71
- Nayak V, Zhao K, Wyce A, Schwartz MF, Lo WS, Berger SL, Marmorstein R* (2006) Structure and dimerization of the kinase domain from yeast Snf1, a member of the Snf1/AMPK protein family. Structure 14(3): 477-485.
- Lo WS, Gamache ER, Harry KW, Yang D, Pillus L and Berger SL* (2005) Activator-targeted Snf1-mediated histone H3 phosphorylation promotes TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24:997-1008
- Lo WS, Henry KW, Schwartz MF and Berger SL* (2004) Histone modification patterns during gene activation. Method in Enzymology 377:130-153
- Clements A, Poux AN, Lo WS, Pillus L, Berger SL, and Marmorstein R* (2003) Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase. Molecular Cell, 12(2):461-473
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NCT, Kao CF, Pillus L, Osley MA and Berger SL* (2003) H2B ubiquitylation and deubiquitylation through the SAGA-associated hydrolase Ubp8 are both required for transcriptional activation. Gene & Dev., 17(21):2648-2663
- Lo WS, Duggan LJ, Belotserkovskya R, Emre NCT, Lane W, Shiekhattar R, and Berger SL* (2001) Snf1 is a histone kinase which works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science, 293:1142-1146.
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R and Berger SL* (2000) Phosphorylation of Serine 10 in Histone H3 Is Functionally Linked In Vitro and In Vivo to Gcn5-Mediated Acetylation at Lysine 14. Molecular Cell 5(6): 917-926.

