
Hsu, Chuan-Chih (許全智)
Assistant Research Specialist
- Ph.D., Biochemistry, Purdue University, USA
- Proteomics Core Laboratory
- cchsu@gate.sinica.edu.tw
- cchsu@as.edu.tw
- +886-2-2787-1157 (Office: A227, A226)
- Academia Sinica Archive
- ORCID
- Web of Science (WOS)
- Google Scholar
The Proteomics Core Lab is dedicated to elucidating the role of protein post-translational modifications (PTMs) in plant cellular mechanisms, focusing on protein folding and enzyme activation processes. Two essential PTMs are N-glycosylation and phosphorylation, which are critical regulators of plant growth and stress responses. Despite their significance, the intricate interplay between two PTMs and their combined effects on kinase activity and signaling pathways is not fully understood. Mass spectrometry (MS) has emerged as a powerful tool for the systematic study of phosphorylation and N-glycosylation, enabling the identification of modification sites and the characterization of N-glycan structures. However, the direct MS detection of these modifications is impeded by challenges such as the low abundance of modified peptides and the heterogeneity of N-glycosylation.
Our lab's research focuses on developing advanced proteomic methodologies to decode phosphorylation- and N-glycosylation-dependent signaling pathways in plants under environmental stress conditions. We specialize in refining sample preparation protocols to enrich phosphorylated and N-glycosylated peptides from plant tissues. Recent advancements include:
1.Suspension Trapping-Based Workflow for In-Depth Plant Phosphoproteomics:
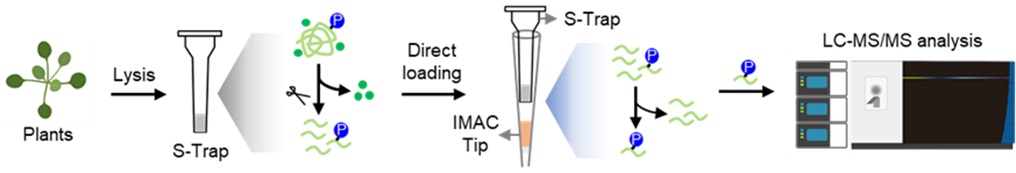
We have pioneered a suspension trapping (S-Trap) based sample preparation workflow, which replaces traditional protein precipitation-based workflow by a tandem S-Trap-IMAC (immobilized metal ion affinity chromatography) approach. This method integrates an S-Trap micro column with an Fe-IMAC tip, significantly enhancing the sensitivity and throughput of plant phosphoproteomics. Our comparative studies show that this strategy increases Arabidopsis protein coverage by over 30%. Utilizing tandem S-Trap-IMAC has provided novel insights into ABA signaling, revealing a prominent role for multiply phosphorylated peptides in early ABA response mechanisms.
2.Streamlined Tandem IMAC-HILIC Workflow for Plant Phosphoproteomics and N-glycoproteomics:
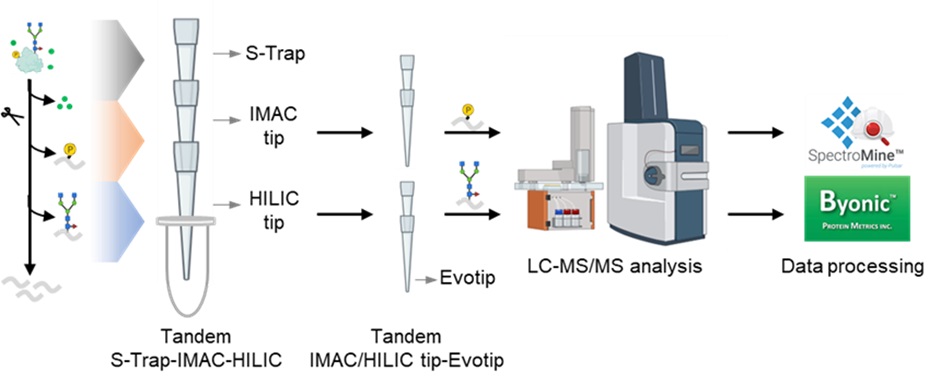
The simultaneous analysis of the phosphoproteome and N-glycoproteome, critical in understanding plant responses to environmental stress, is often hindered by laborious and time-consuming enrichment methods. We addressed this bottleneck by developing a novel tandem S-Trap-IMAC-HILIC (TIMAHAC) strategy. TIMAHAC integrates IMAC and HILIC enrichments into a tandem tip format, utilizing a unified buffer system and optimized enrichment sequence to improve efficiency and reproducibility. Our methodology was validated by analyzing the Arabidopsis phosphoproteome and N-glycoproteome following ABA treatment, identifying up to 1,954 N-glycopeptides and 11,255 phosphopeptides. TIMAHAC demonstrates its potential for high-throughput and simultaneous analysis, offering a comprehensive solution for exploring the complex PTM landscape and regulatory networks in plant biology.
- Sang, T.; Chen, C.-W.; Lin, Z.; Ma, Y.; Du, Y.; Lin, P.-Y.; Hadisurya, M.; Zhu, J.-K.; Lang, Z.; Tao, W. A; Hsu, C.-C.*; Wang, P.* DIA-based phosphoproteomics identifies early phosphorylation events in response to EGTA and mannitol in Arabidopsis. Mol. Cell. Proteomics 2024, 23, 100804.
- Chen, C.-W.; Lin, P.-Y.; Lai, Y.-M.; Lin, M.-H.; Lin, S.-Y.; Hsu, C.-C.* TIMAHAC: streamlined tandem IMAC-HILIC workflow for simultaneous and high-throughput plant phosphoproteomics and N-glycoproteomics. Mol. Cell. Proteomics 2024, 23, 100762.
- Lin, Y.-H.#; Xu, M.-Y.#; Hsu, C.-C. #; Lee, H.-C.; Damei, F. A.; Lee, H.-C.; Tsai, W.-L.; Hoang, C. V.; Chiang, Y.-R.; Ma, L.-S. Ustilago maydis PR-1-like protein has evolved two distinct domains for dual virulence activities. Nat. Commun. 2023, 14, 5755. #equal contribution.
- Chen, C.-W.; Tsai, C.-F.; Lin, M.-H.; Lin, S.-Y.; Hsu, C.-C.* Suspension trapping-based sample preparation workflow for sensitive plant phosphoproteomics. Anal. Chem., 2023, 95, 12232-12239.
- Kim, T.-W.#; Park, C. H.#; Hsu, C.-C.#; Kim, Y.-W.; Ko, Y.-W.; Zhang, Z.; Zhu, J.-Y.; Hsiao, Y.; Branon, T.; Kaasik, K.; Saldivar, E.; Li, K.; Pasha, A.; Provart, N. J.; Burlingame, A. L.; Xu, S.-L.; Ting, A. Y.; Wang, Z.-Y. Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics. Plant Cell 2023, 35, 975-993. #equal contribution.

Lab Members
陳瑾玟 Chin-Wen Chen
助理專案研發學者 Assistant R&D Scientist (2008.09-)
林佩怡 Pei-Yi Lin
副專案研發學者 Associated R&D Scientist (2022.11-)
陳亭安 Ting-An Chen
研究助理 Research Assistant (2024.02-)
陳柏蒼 Po-Tsang Chen
研究助理 Research Assistant (2025.09-) (農生)
Former Members
蕭証元 Jeng-Yuan Shiau
研究助理 Research Assistant (農生)
連雅苹 Ya-Ping Lien
研究助理 Research Assistant
謝豐如 Feng-Ju Hsieh
研究助理 Research Assistant (農生)
葉家豪 Chia-Hao Yeh
研究助理 Research Assistant (農生)
嚴孝榕 Hsiao-Jung Yen
研究助理Research Assistant (2014.10-2020.12) (農生)
賴映糸 Ying-Mi Lai
研究助理Research Assistant (2021.03-2023.05) (農生)
鄧怡君 Yi-Chun Teng
研究助理Research Assistant (2023.11-2024.02) (農生)
Domestic
- 2024: Technician Professional Award, Taiwan Society for Mass Spectromrtry