
Wu, Shu-Hsing (吳素幸)
Distinguished Research Fellow
- Ph.D., Plant Biology, University of California, Davis, USA
- Light-mediated gene expression and signal transduction in Arabidopsis
- shuwu@gate.sinica.edu.tw
- shuwu@as.edu.tw
- +886-2-2787-1066 (Lab: A326)
- +886-2-2787-1178 (Office: A328)
- Academia Sinica Archive
- ORCID
- Web of Science (WOS)
- Google Scholar
Light-mediated gene expression and signal transduction in Arabidopsis
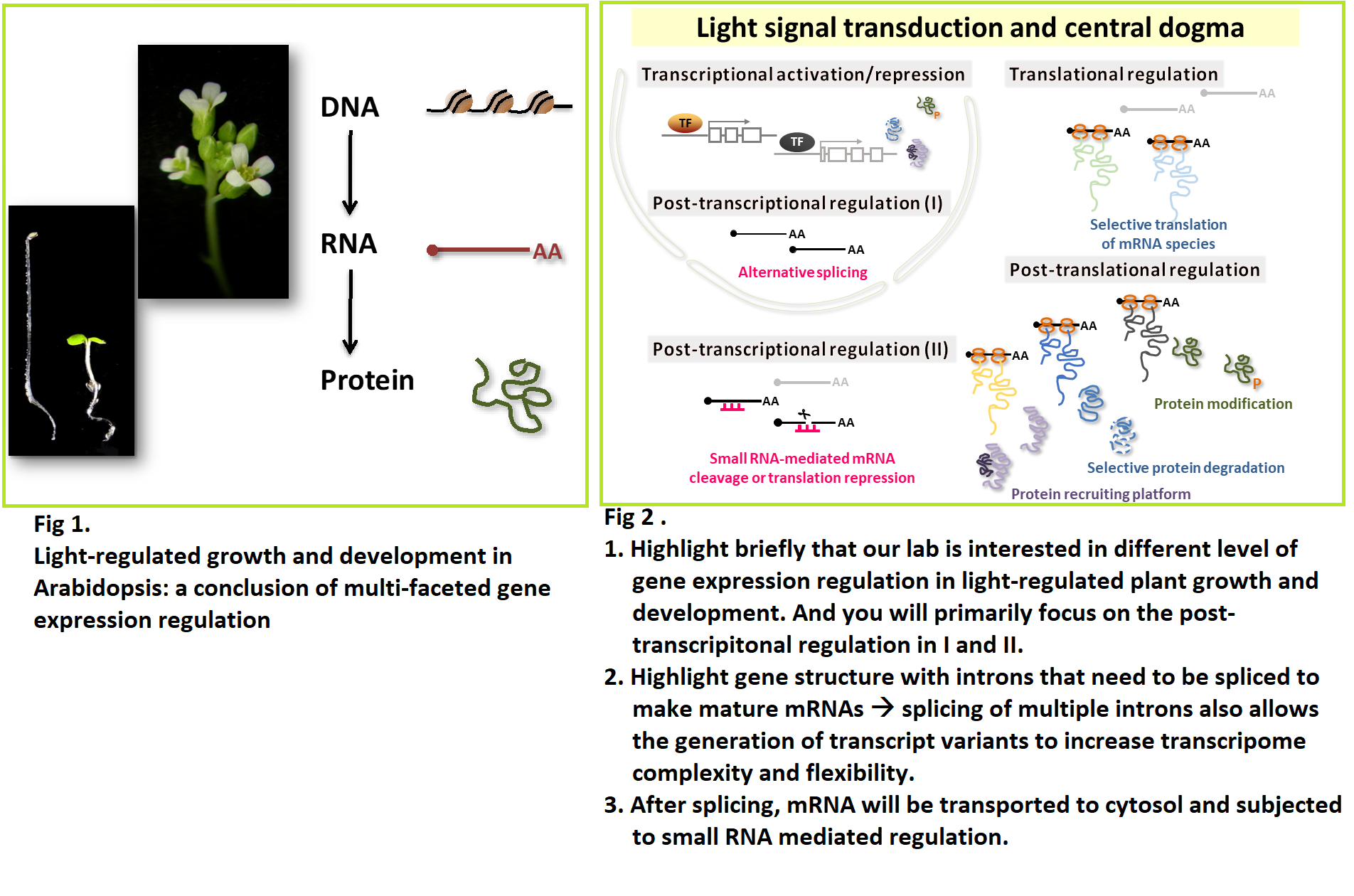
“Light” regulates multiple aspects of growth and development in plants. Plants have a collection of photoreceptors to interpret and transduce the light signals. The light-mediated morphogenesis is the conclusion of gene expression and interactions. Light influences gene expression at multiple levels, from transcription to post-translational modification. When the expression and interactions of genes are harmonically coordinated, plants achieve optimal growth and development under ever-changing light environment. Our primary goal is to identify genes and regulatory mechanisms for gene expression associated with light-regulated plant growth and development.
Our studies have revealed that, in addition to the commonly investigated transcriptional regulation and selective protein degradation, post-transcriptional regulation plays an important role in Arabidopsis photomorphogenic development. Small RNAs function in maintaining adequate mRNA abundance in cells and alternative splicing increases transcript and proteome diversities. We have also observed a clear impact of light on selective protein translation.
Light also sets the circadian clock in plants for them to coordinate internal biochemical processes for the anticipation of the 24-h light-dark cycles. The circadian clock in Arabidopsis consists of several interlocked feedback loops, yet additional regulatory components remain to be discovered. We found that LWD1 and LWD2 are two clock proteins essential for the stabilization and robustness of Arabidopsis circadian clock. In addition to be part of a positive feedback loop in Arabidopsis circadian clock, LWD also functions as a co-activator via the interaction with the TCP transcription factor to activate the expression of the clock morning gene CCA1 at dawn.
Our laboratory will continue to discover hidden components in light signaling pathways. We will also dedicate our efforts in decoding the action mechanisms of the components identified.
- Wu, CC, Larsson L, Yu CP, Chen YH, Ding KH, Yang HC, Lundeberg J, Ho CM*, Wu SH*, Lu MY* and Li WH* (2026) Serial spatial transcriptomes reveal regulatory transitions in maize leaf development. Plant Biotech. J. (accepted)
- Lu KJ, Hsu CW, Jane WN, Peng MH, Chou YW, Huang PH, Yeh KC, Wu SH and Liao JC* (2025) Dual-cycle CO2 fixation enhances growth and lipid synthesis in Arabidopsis thaliana. Science, 389(6765): p. eadp3528.
- Huang CF, Liu WY, Yu CP, Wu SH, Ku MSB* and Li WH* (2023) C4 leaf development and evolution. Curr. Opin. Plant Biol. 76:102454
- Wu HW, Fajiculay E, Wu JF, Yan CCS, Hsu CP* and Wu SH* (2022) Noise reduction by upstream open reading frames. Nature Plants 8(5):474-480
- Huang CK, Lin WD and Wu SH* (2022) An improved repertoire of splicing variants and their potential roles in Arabidopsis photomorphogenic development. Genome Biol 23:50
- Chen HH, Yu HI, Rudy R, Lim SL, Chen YF, Wu SH, Lin SC, Yang MH and Tarn WY (2021) DDX3 modulates the tumor microenvironment via its role in endoplasmic reticulum-associated translation. iScience 24(9):103086.
- Sanobar N, Lin PC, Pan ZJ, Fang RY, Tjita V, Chen FF, Wang HC Tsai HL, Wu SH, Shen TL, Chen YH and Lin SS* (2021) Investigating the viral suppressor HC-Pro inhibiting small RNA methylation through functional comparison of HEN1 in angiosperm and bryophyte. Viruses 13(9): 1837
- Joanito I, Sanders CC, Chu JW, Wu SH and Hsu CP* (2020) Basal leakage in oscillation: coupled transcriptional and translational control using feed-forward loops. PLOS Computational Biology, in press.
- Jang GJ, Jang JC, Wu SH* (2020) Dynamics and functions of stress granules and processing bodies in plants. Plants 9:1122
- Liu WY, Lin HH, Yu CP, Chang CK, Chen HJ, Lin JJ, Lu MY, Tu, SL, Shiu, SH, Wu SH*, Ku MSB* and Li WH* (2020) Maize ANT1 modulates vascular development, chloroplast development, photosynthesis and plant growth. Proc Natl. Acad. Sci. USA doi/10.1073/pnas.2012245117
- Yu X, Li B, Jang GJ, Jiang S. Jiang D, Jang JC, Wu SH, Shan L and He P* (2019) Orchestration of processing body dynamics and mRNA decay in Arabidopsis immunity. Cell Rep. 28:2194-2205
- Jang GJ, Yang JY, Hsieh HL and Wu SH* (2019) Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc Natl. Acad. Sci. USA 116:6451-6456
- Chang YM, Lin HH, Liu WY, Yu CP, Chen HJ, Wartini PP, Kao YY, Wu YH, Lin JJ, Lu MY, Yu SL, Wu SH, Shiu SH, Ku MSB and Li WH* (2019) A Comparative Transcriptomics Method to Infer Gene Coexpression Networks and its applications to Maize and Rice Leaf Transcriptomes. Proc Natl. Acad. Sci. USA 116:3091-3099
- Chen GH, Liu MJ, Xiang Y, Sheen J and Wu SH* (2018) Target of rapamycin (TOR) and ribosomal protein S6 (RPS6) transmit light signals to enhance protein translation in de-etiolating Arabidopsis seedlings. Proc Natl. Acad. Sci. USA 115:12823-12828
- Joanito I, Chu JW, Wu SH, and Hsu CP* (2018) An incoherent feed-forward loop switches the Arabidopsis clock rapidly between two hysteretic states. Scientific Reports 8:13944
- Huang CF, Yu CP, Wu YH, Lu MJ, Tu SL, Wu SH, Shiu SH, Ku MSB and Li WH* (2017) Elevated auxin biosynthesis and transport underlie high vein density in C4 leaves. Proc Natl. Acad. Sci. USA 114: E6884-E6891
- Lin MC, Tsai HL, Lim SL, Jeng ST and Wu SH* (2017) Unraveling multifaceted contributions of small regulatory RNAs to photomorphogenic development in Arabidopsis. BMC Genomics. 18: 559
- Huang WY, Wu YC, Pu HY, Wang Y, Jang GJ and Wu SH* (2017) Plant dual-specificity tyrosine phosphorylation-regulated kinase optimizes light-regulated growth and development in Arabidopsis. Plant Cell Environ. 40: 1735-1747
- Li C, Sako Y, Imai A, Nishiyama T, Thompson K, Kubo M, Hiwatashi Y, Kabeya Y, Karlson D, Wu SH, Ishikawa M, Murata T, Benfey P.N, Sato Y, Tamada Y, Hasebe M* (2017) A Lin28 homologue reprograms differentiated cells to stem cells in the moss Physcomitrella patens. Nat. Commun. 8: 14242
- Wu J.-F., Tsai H.-L., Joanito I., Wu Y.-C., Chang C.-W., Li Y.-H., Wang Y., Hong J. C., Chu J.-W., Hsu C.-P., and Wu S.-H. (2016) LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 7, 13181 doi: 10.1038/ncomms13181
- 2.Huang C.-F., Chang Y.-M., Lin J.-J., Yu C.-P., Lin H.-H., Liu W.-Y., Yeh S., Tu S.-L., Wu S.-H., Ku M. S.-B.*, and Li W.-H.* (2016) Insights into the regulation of C4 leaf development from comparative transcriptomic analysis. Curr. Opin. Plant Biol. 30: 1-10
- Lee W.-C., Lu S.-H., Lu M.-H., Yang C.-J., Wu S.-H., Chen H.-M* (2015) Asymmetric bulges and mismatches determine 20-nucleotide microRNA formation in plants. RNA Biol. 12: 1054-66
- Yu C.-P., Chen S. C.-C., Chang Y.-M., Liu W.-Y., Lin H.-H., Lin J.-J., Chen H.-J., Lu Y.-J., Wu Y.-H., Lu M.-Y. J., Lu C.-H., Shih A. C.-C., Ku M. S.-B., Shiu S.-H.*, Wu S.-H.*, Li W.-H.* (2015) Transcriptome dynamics of developing maize leaves and genome-wide prediction of cis elements and their cognate transcription factors. Proc. Natl. Acad. Sci. USA 112: E2477-86
- Tsai H.-L., Li Y.-H., Hsieh W.-P., Lin M.-C., Ahn J. H., Wu S.-H.* (2014) HUA ENHANCER 1 is involved in post-transcriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell 26: 2858-2872
- Wu H.-Y., Liu K.-H., Wang Y.-C., Wu J.-F., Chiu W.-L., Chen C.-Y., Wu S.-H., Sheen J., Lai E.-M.* (2014) AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 10:19
- Wu S.-H.* (2014) Gene expression regulation in photomorphogenesis: from the perspective of central dogma. Ann. Rev. Plant Biol. 65:311-333
- Liu M.-J., Wu S.-H., Wu J.-F., Lin W.-D., Wu Y.-C., Tsai T.-Y, Tsai H.-L., Wu S.-H.* (2013) Translational landscape of photomorphogenic Arabidopsis. Plant Cell 10: 3699-710
- Chen C.-E., Yeh K.-C., Wu S.-H., Wang H.-I., Yeh H.-H.* (2013) A vicilin-like seed storage protein, PAP85, is involved in tobacco mosaic virus replication. J Virol. 87:6888-68900
- Chen YY, Wang Y, Shin LJ, Wu JF, Shanmugam V, Tsednee M, Lo JC, Chen CC, Wu SH, and Yeh KC.* (2013) Iron is involved in maintenance of circadian period length in Arabidopsis. Plant Physiol. 161:1409-1420.
- Hsieh WP, Hsieh HL, Wu SH.* (2012) Arabidopsis bZIP16 transcription factor integrates light and hormone signaling pathways to regulate early seedling development. Plant Cell 24: 3997–4011
- Liu MJ, Wu S, Chen HM, Wu SH.* (2012) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Molecular Systems Biology 8:566.
- Chang CS, Maloof JN, Wu SH.* (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156:228-239.
- Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, and Wu SH.* (2011) LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23: 486–498.
- Chen HM, Chen LT, Patel K, Li YH., Baulcombe D, and Wu SH* (2010) 22-nucleotide RNA triggers secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107:15269-15274.
- Khanna R*, Kronmiller BA, Maszle DR, Coupland G, Holm M, Mizuno T, and Wu SH. (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21:3416-3420.
- Wu JF, Wang Y, and Wu SH.* (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 148:948-959
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, Somerville S, and Wu SH.* (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54:205-219.
- Chen HM, Li YH, and Wu SH.* (2007) Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104:3318-3323.


吳靜芬 Jing-Fen Wu Associate R&D Scientist, 副專案研發學者

吳怡蓁 Yi-Chen Wu Associate R&D Scientist, 副專案研發學者 wyc9823@gate.sinica.edu.tw

黃俊凱 Chun-Kai Huang R&D Scientist, 專案研發學者 hck0903@gate.sinica.edu.tw

卓岳 Yueh Cho Postdoctoral Scholar, 博士後研究學者 choyueh@gate.sinica.edu.tw

林岑穎 Tsen-Ying Lin Research Assistant 研究助理 crosswing052@gmail.com

陳彥麇 Yen-Chiun Chen TIGP-MBAS 博士班學生 PhD student wotan13233@gmail.com

潘慕得 Mu-Te Pan NTU 博士班學生 PhD student mandypan112385@gmail.com

凌郁婷 Yu-Ting Ling Research Assistant 研究助理 as0210027@gate.sinica.edu.tw

鄧佩珊 Phoi San Tran Research Assistant 研究助理 phoisantran1993@gate.sinica.edu.tw

楊宜蓉 Yi-Rong Yang Research Assistant 研究助理 yangyirong12@gate.sinica.edu.tw

殷筱婷 Siao-Ting Yan Administrative Assistant 行政助理 yst1226@gate.sinica.edu.tw
International
- 2021: Enid MacRobbie Corresponding Membership Award, American Society of Plant Biologists, USA
- 2015: Asia and Oceania Society for Photobiology (AOSP) Award, AOSP
- 2006: Thomson Citation Laureate Award, Thomson Reuters Corporation
Domestic
- 2024: Frontier Science Research Program, National Science and Technology Council
- 2023: Investigator Award, Academia Sinica
- 2019: Frontier Science Research Program, Ministry of Science and Technology
- 2015: The 59th Academic Awards, Ministry of Education
- 2015: Outstanding Research Award, Ministry of Science and Technology
- 2014: Frontier Science Research Program, Ministry of Science and Technology
- 2013: Investigator Award, Academia Sinica
- 2012: Outstanding Research Award, National Science Council
- 2010: Shang-Fa Yang Young Scientist Award, The Shang-Fa Yang Memorial Foundation
- 2008: Junior Research Investigator Award, Academia Sinica