
Ma, Ka-Wai (馬家威)
Assistant Research Fellow
- Ph.D., Plant Pathology, University of California, Riverside, USA
- Plant innate immunity, molecular mechanisms regulating plant-microbiota homeostasis
- kawaim7@gate.sinica.edu.tw
- kawaim7@as.edu.tw
- +886-2-2787-1060 (Lab: R326)
- +886-2-2787-1100 (Office: R326)
- X:@KaWaiMa6
- ORCID
- Web of Science (WOS)
- Google Scholar
The Ma Lab
The Institute of Plant and Microbial Biology (IPMB), Academia Sinica, Taiwan
The Microbiome
Higher-level organisms, including terrestrial plants, have associations with multifaceted microbial communities known as microbiota/microbiome. Plants invest up to 11% of their carbon that is derived from photosynthesis as root exudate and rhizodeposition to attract and nourish their associated microbiota. Plants benefit from microbial services including boosted protection against disease, improved tolerance to stress, and enhanced nutrient uptake. However, in order to limit pathogen overgrowth, plants must also develop strong immunity. Plant physiology and microbiota activities are thus interdependent and function as a single biological unit referred to as the Holobiont. Commensalism (o,+), pathogenesis (-,+), competition (-,-) and mutualism (+,+) are the broad categories that pairwise plant-microbe interactions fall into. These interactions are not often exclusive and can change dynamically in response to alterations in the host and environment. The Ma lab strives to understand various principles and mechanisms that regulate shifting of these interactions.
Current projects include the following
- Understanding the molecular mechanisms of how commensal bacteria interferes with plant immune responses.
- Employing multi-omics and genetics to identify plant and microbial genetic determinants that influence holobiont interactions.
- Formulating microbiota-based products to enhance plant fitness.
The Ma Lab will relocate to the Institute of Plant and Microbial Biology (IPMB), Taiwan, in December 2023. We invite applications from enthusiastic students and postdoctoral researchers interested in plant-microbiota interaction and plant pathology. Please get in touch!
- Understanding the molecular mechanisms of how commensal bacteria interferes with plant immune responses.
Our previous work has shown that bacterial commensals from major taxonomic group can interfere with the plant immune responses. We are interested in understanding
I) the molecular mechanisms of interference, and
II) how interference with plant immunity contributes to altered microbiota establishment
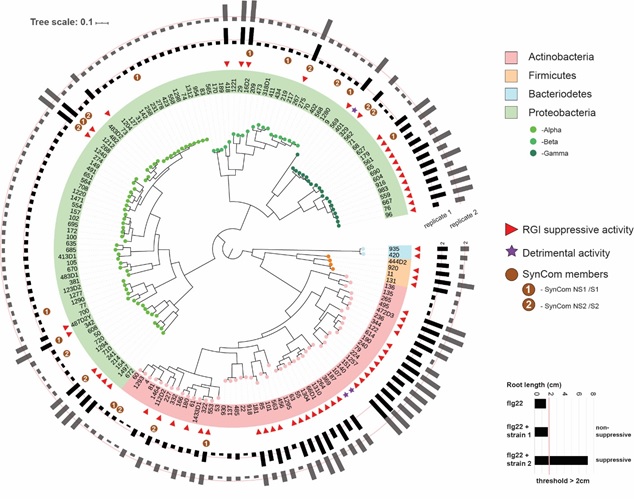
Modified from Ma et al., Nature Plants 2021. Phylogenetic tree indicating the strain-specific variation of At-R-sphere bacterial microbiota members to interfere with plant immune response.
- Employing multi-omics and genetics to identify plant and microbial genetic determinants that influence holobiont interactions.
Plants colonized by its associated microbiota express dramatically different transcriptomics compared to axenic plants. By inoculating plants with microbes/microbial community of specific traits, we can narrow down candidate genes that are commonly or specifically regulated in response to the microbiota. These genes are candidates for functional studies and targets of plant traits amelioration. Currently, we are focusing on genes that are differentially regulated by microbes with or without the capacity to interfere with plant immune responses.
- Formulating microbiota-based products to enhance plant fitness.
Although plant breeding and genetic engineering (GE) approaches can improve plant fitness by modifying plant genotypes, this is usually time-consuming and may be limited by regional GE regulations. Manipulating the microbiota therefore offers a rapid and GM-free alternative to improve plant traits.
As a proof of principle, my previous study showed that two plant traits, defence and growth, are coordinated by the interaction between the plant host and its associated microbiota with contrasting traits to interfere with the host immune response. Such a delicate balance of plant-microbe interactions acts analogously as a rheostat to maintain homeostasis. We aim to understand the principles of plant microbiota assembly and design simplified microbial communities as formulations to enhance plant fitness.
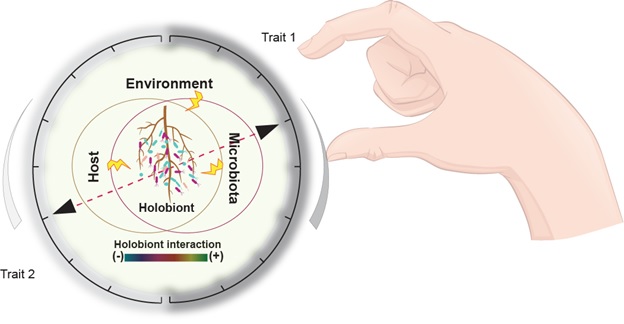
A schematic model illustrating how plant microbiota homeostasis could be manipulated to affect plant traits of interest.
All publications
Selected Publications
# First authors
* Corresponding author
- C. Copeland, P. Schulze-Lefert, K.-W. Ma *, Potential and challenges for application of microbiomes in agriculture. Plant Cell 37, koaf185 (2025).
- K.-W. Ma*, Coordinated action by individuals orchestrates infection through the division of labour. Nat. Rev. Microbiol. 23, 549–549 (2025).
- T.-T. Lu, M. Isip, H.-J. S. Shih, S. Perin, K.-W. Ma*, Misregulation of the jasmonate signaling pathway leads to altered plant microbiota interaction and plant stress responses. bioRxiv [Preprint] (2025). https://doi.org/10.1101/2025.03.29.646076.
- J. Ordon, E. Logemann, L.-P. Maier, T. Lee, E. Dahms, A. Oosterwijk, J. Flores-Uribe, S. Miyauchi, L. Paoli, S. C. Stolze, H. Nakagami, G. Felix, R. Garrido-Oter, K.-W. Ma *, P. Schulze-Lefert*, Conserved immunomodulation and variation in host association by Xanthomonadales commensals in Arabidopsis root microbiota. Nat. Plants 11, 612–631 (2025).
- B. N. Kunkel, J. Ludwig-Müller, K.-W. Ma, Editorial: The role of auxin in plant-microbe interactions. Front. Plant Sci. 15 (2024).
- J. Ordon, J. Thouin, R. T. Nakano, K.-W. Ma, P. Zhang, B. Huettel, R. Garrido-Oter, P. Schulze-Lefert, Chromosomal barcodes for simultaneous tracking of near-isogenic bacterial strains in plant microbiota. Nat. Microbiol. 9, 1117–1129 (2024).
- F. Getzke, M. A. Hassani, M. Crüsemann, M. Malisic, P. Zhang, Y. Ishigaki, N. Böhringer, A. Jiménez Fernández, L. Wang, J. Ordon, K.-W. Ma, T. Thiergart, C. J. Harbort, H. Wesseler, S. Miyauchi, R. Garrido-Oter, K. Shirasu, T. F. Schäberle, S. Hacquard, P. Schulze-Lefert, Cofunctioning of bacterial exometabolites drives root microbiota establishment. Proc. Natl. Acad. Sci. 120, e2221508120 (2023).
- K.-W. Ma#, J. Ordon, P. Schulze-Lefert, Gnotobiotic Plant Systems for Reconstitution and Functional Studies of the Root Microbiota. Curr. Protoc. 2, e362 (2022).
- K.-W. Ma#, Y. Niu#, Y. Jia#, J. Ordon, C. Copeland, A. Emonet, N. Geldner, R. Guan, S. C. Stolze, H. Nakagami, R. Garrido-Oter, P. Schulze-Lefert, Coordination of microbe–host homeostasis by crosstalk with plant innate immunity. Nat. Plants 7, 814–825 (2021).
- A. Emonet, F. Zhou, J. Vacheron, C. M. Heiman, V. Dénervaud Tendon, K.-W. Ma, P. Schulze-Lefert, C. Keel, N. Geldner, Spatially Restricted Immune Responses Are Required for Maintaining Root Meristematic Activity upon Detection of Bacteria. Curr. Biol. 31, 1012-1028.e7 (2021).
- R. Garrido-Oter, R. T. Nakano, N. Dombrowski, K.-W. Ma, A. C. McHardy, P. Schulze-Lefert, Modular Traits of the Rhizobiales Root Microbiota and Their Evolutionary Relationship with Symbiotic Rhizobia. Cell Host Microbe 24, 155-167.e5 (2018).
- Z.-M. Zhang#, K.-W. Ma#, L. Gao, Z. Hu, S. Schwizer, W. Ma, J. Song, Mechanism of host substrate acetylation by a YopJ family effector. Nat. Plants 3, 1–10 (2017).
- Z.-M. Zhang#, K.-W. Ma#, S. Yuan, Y. Luo, S. Jiang, E. Hawara, S. Pan, W. Ma, J. Song, Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat. Struct. Mol. Biol. 23, 847–852 (2016).
- K.-W. Ma#, W. Ma, YopJ Family Effectors Promote Bacterial Infection through a Unique Acetyltransferase Activity. Microbiol. Mol. Biol. Rev. 80, 1011–1027 (2016).
- K.-W. Ma#, W. Ma, Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol. Biol. 91, 713–725 (2016).
- K.-W. Ma#, S. Jiang, E. Hawara, D. Lee, S. Pan, G. Coaker, J. Song, W. Ma, Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 208, 1157–1168 (2015).
- S. Jiang, J. Yao, K.-W. Ma, H. Zhou, J. Song, S. Y. He, W. Ma, Bacterial Effector Activates Jasmonate Signaling by Directly Targeting JAZ Transcriptional Repressors. PLOS Pathog. 9, e1003715 (2013).
- K.-W. Ma#, C. Flores, W. Ma, Chromatin Configuration as a Battlefield in Plant-Bacteria Interactions. Plant Physiol. 157, 535–543 (2011).
