Hsieh, Phoebe Yu-Ying (謝侑穎)
Assistant Research Fellow
- Molecular and Cellular Biology, Harvard University, USA
- Polymicrobial interaction, Microbial ecology and evolution, Genetics and functional genomics, Experimental evolution
- yhsieh@fredhutch.org
- yhsieh@as.edu.tw
- +886-2-2787-1113 (Office: A330/R407 (temporary Office))
- Lab Website
- ORCID
- Google Scholar

Microbes—fungi, bacteria, and phages—rarely live in isolation. They constantly interact, cooperating or competing in diverse niches, like soil or within plant and animal hosts. Microbial competition is especially intense in polymicrobial infections, where multiple microbes co-colonize the same host and must contend with nutrient limitation and host immune responses
How these microbial battles unfold and how they drive microbial evolution remain poorly understood. At the lab of PIE, we study polymicrobial interactions using fungi and bacteria that cause infectious diseases in plants or humans. We aim to understand the mechanisms and consequences of polymicrobial competition and how these interactions shape host-microbe dynamics.
Using bottom-up approaches — including functional genomics and experimental evolution — we study how bacterial pathogens, like Pseudomonas aeruginosa (in humans) and Pseudomonas syringae (in plants), engage with fungal and phage rivals, in both in vitro and in vivo models. Through these models, we aim to uncover fundamental principles of microbial competition and adaptations to complex biotic pressures. Our long-term goal is to harness this knowledge to engineer microbial communities or develop effective antimicrobial strategies for medical and agricultural purposes.
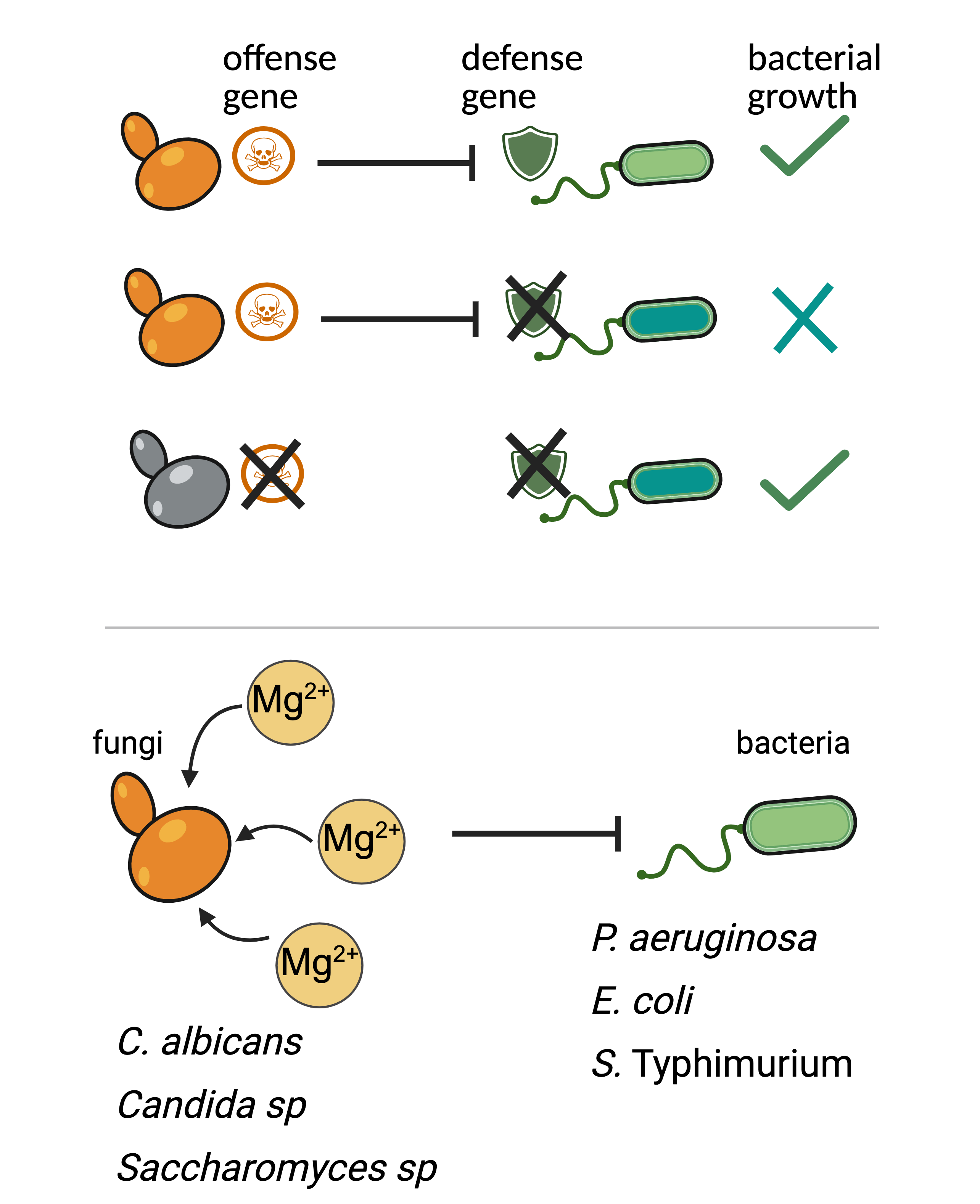
Genetic conflicts in fungal-bacterial competition
Competition is inevitable in polymicrobial communities, where microbes share similar niches and must evolve strategies to secure resources or space. While interbacterial competition is well studied—often involving toxins or nutrient sequestration—less is known about how bacteria compete with fungi. Do bacteria use different molecular arsenals against fungal competitors? And how can we identify these mechanisms systematically?
We view microbial competition as a genetic conflict, where one species expresses an "offense" gene to antagonize another, which must protect itself either by evasion or by expressing a "defense" gene. Using this framework and a genome-wide bacterial fitness screen, we previously discovered that the human fungal pathogen Candida albicans sequesters Mg2+ to suppress the growth of the human bacterial pathogen Pseudomonas aeruginosa. This nutritional competition for Mg2+ is a widespread mode of competition between multiple fungi and gram-negative bacteria.
Building on this foundation, we are interested in understanding the mechanisms through which other Pseudomonas species engage in fungal competition, how diverse fungal species mount defense, and how bacteria and fungi adapt in this arms race.

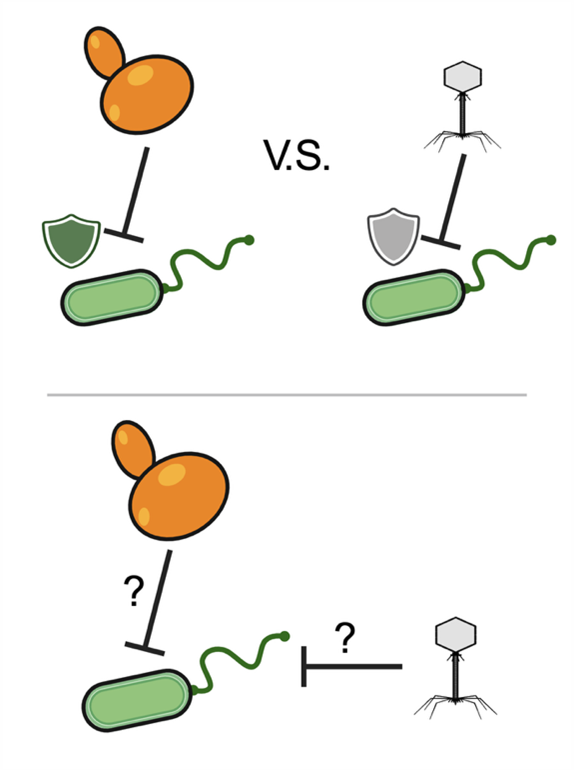
Genetic conflicts in fungal-bacterial-phage interactions
In polymicrobial communities, bacteria must compete not only with neighboring fungi but also defend themselves against viral predators—bacteriophages. Phages hijack bacterial cellular machinery to replicate; in response, bacteria evolve defenses that block phage entry or replication. At the same time, bacteria are likely to engage in resource competition and antagonism with fungi.
This raises key questions: How do bacteria manage simultaneous selective pressures from both fungi and phage? Does competition with fungi alter bacterial susceptibility to phages, or vice versa? We are interested in understanding the genes bacteria use to defend against fungi, phages, or both, and how these cross-kingdom conflicts jointly drive bacterial evolution.
Polymicrobial interactions at the host-microbe interface
Our previous work revealed that fungal-bacterial competition for Mg2+ alters bacterial adaptation to antibiotics. This indicates that competition may broadly impact how microbes respond or adapt to other stressors — such as host immune defense during infection.
To explore this further, we use plant and animal infection models combined with microbial genetics and genomics, and cell biology experiments. We aim to identify fungal and bacterial genes critical for polymicrobial infections and to understand how cross-kingdom microbial interactions drive the progression of infectious diseases and host response. Ultimately, our goal is to understand how these interactions shape microbial virulence and adaptation in complex, host-associated ecosystems.
Interested in joining us? Contact Phoebe to discuss further project ideas
All publications
Selected Publications
For a full list of Phoebe's publications, click here
- Hsieh, Y. -Y.+*,O’Keefe+, I., Sun, W., Wang, Z. C., Vu, L., Ernst, R., Dandekar, A. A., Malik, H. S. (2024) “Magnesium depletion unleashes two unusual modes of colistin resistance with different fitness costs” BioRxiv preprint DOI: 10.1101/2024.10.15.618514
- Hsieh, Y. -Y.*, Sun, W., Young, J. M., Cheung R., Hogan, D. A., Dandekar, A. A., Malik, H. S.* (2024) “Widespread fungal-bacterial competition for magnesium lowers bacterial susceptibility to polymyxin antibiotics” PLOS BiologyDOI: 10.1371/journal.pbio.3002694
- Hsieh, Y. -Y.*, Makrantoni, V., Robertson D., Marston, A. L., Murray, A. W.* (2020) “Evolutionary repair: changes in multiple functional modules allow meiotic cohesin to support mitosis”
- LaBar T., Hsieh, Y. -Y., Fumasoni M., Murray, A. W.* (2020) “Evolutionary repair experiments as a window to the molecular diversity” Current Biology30: R565-R574 DOI: 10.1016/j.cub.2020.03.046
- Hsieh, Y.-Y., Hung, P.-H., Leu, J-Y.* (2013) “Hsp90 regulates non-genetic variation in response to environmental stress” Molecular Cell50: 82–92 DOI: 10.1016/j.molcel.2013.01.026
- McDonald, M.J., Hsieh, Y.-Y., Yu, Y.-H., Chang, S.-L. and Leu, J.-Y.* (2012) “The evolution of low mutation rates in experimental mutator populations of Saccharomyces cerevisiae” Current Biology22: 1235–1240 DOI: 10.1016/j.cub.2012.04.056