
Hsieh, Ming-Hsiun (謝明勳)
Research Fellow
- Ph.D., Biology, New York University, USA
- Plant molecular nutrition and signal transduction
- ming@gate.sinica.edu.tw
- ming@as.edu.tw
- +886-2-2787-1046 (Lab: A420)
- +886-2-2787-1168 (Office: A440)
- Academia Sinica Archive
- ORCID
- Google Scholar
Ming-Hsiun Hsieh studied nitrogen (N) sensing and metabolism in the laboratory of Gloria M. Coruzzi, where he worked on the N regulatory protein PII, glutamate receptor-like proteins (GLRs), and asparagine synthetase in Arabidopsis. He discovered genes encoding ACT domain repeat (ACR) and heptahelical transmembrane protein (HHP) families and investigated the methylerythritol phosphate pathway mutants in Arabidopsis under the supervision of Howard M. Goodman. The Hsieh laboratory at Academia Sinica has contributed to the research of RNA metabolism in chloroplasts and mitochondria, vitamin B1 biosynthesis, and glutamine (Gln)/glutamate (Glu) metabolism and signaling in plants.
Gln/Glu signaling in Arabidopsis
Gln and Glu are the first organic nitrogen synthesized in the plant cell. In addition to their role in metabolism, we recently found that Gln and Glu could activate the expression of stress- and defense-responsive genes in plants. We propose a working model in which extracellular Gln and Glu may serve as “danger signals” to activate plant defense (Fig. 1).

Fig. 1 A hypothetical working model for Gln/Glu functions in plant growth and defense. GS, Gln synthetase; GOGAT, Gln:2-oxoglutarate aminotransferase; ACR11, ACT domain repeat 11.
The ACR proteins are putative amino acid sensors in plants. Arabidopsis has twelve ACRs, and their functions are largely unknown. The ACR11 protein interacts and regulates Gln synthetase 2 (GS2) and Fd-dependent Gln:2-oxoglutarate aminotransferase 1 (Fd-GOGAT1), but the molecular mechanisms remain elusive. We are interested in studying the functions of Arabidopsis ACRs, including the mechanism of ACR11 in regulating GS2/Fd-GOGAT1 (Fig. 1).
To study genes involved in Gln/Glu-regulated growth and defense, we screened for Arabidopsis mutants that grew poorly on Gln or Glu as the sole N source (Fig. 2). We are in the process of identifying the mutated genes and the underlying molecular mechanisms.
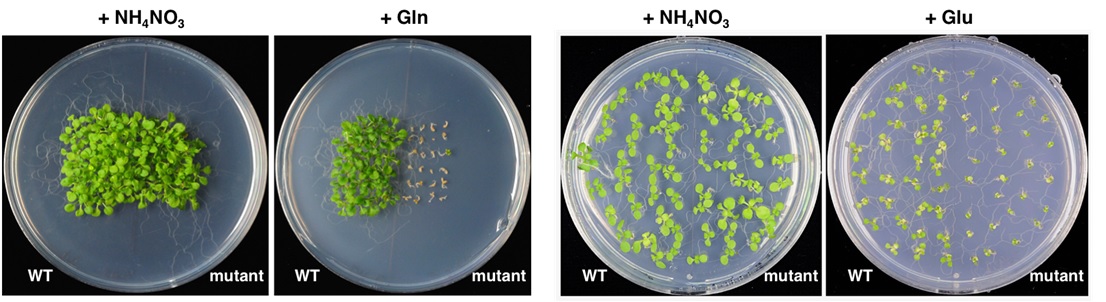
Fig. 2 Arabidopsis mutants grown on medium containing Gln or Glu as the sole nitrogen source.
Gln/Glu signaling in rice
We previously found that Gln and Glu could rapidly induce the expression of stress-responsive genes in rice (Oryza sativa). To study the functions of NH4NO3-, Gln-, and Glu-responsive genes, we use the CRISPR/Cas9 technology to generate a collection of rice mutants (Fig. 3). Further study on the CRISPR mutants may provide insights into the functions of these genes and the molecular mechanism of Gln/Glu-regulated stress response in rice.
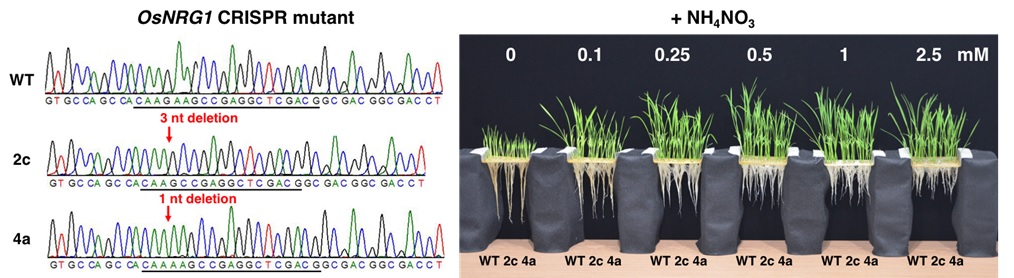
Fig. 3 Genotypes and phenotypes of the OsNRG1 CRISPR mutants.
- Liao HS, Lee KT, Chung YH, Chen SZ, Hung YJ, Hsieh MH* (2024) Glutamine induces lateral root initiation, stress responses, and disease resistance in Arabidopsis. Plant Physiol (accepted)
- Chung YH, Chen TC, Yang WJ, Chen SZ, Chang JM, Hsieh WY, Hsieh MH* (2024) Ectopic expression of a bacterial thiamin monophosphate kinase enhances vitamin B1 biosynthesis in plants. Plant J 117: 1330–1343
- Lee KT, Liao HS, Hsieh MH* (2023) Glutamine metabolism, sensing, and signaling in plants. Plant Cell Physiol 64: 1466–1481
- Liao HS, Chen YJ, Hsieh WY, Li YC, Hsieh MH* (2023) Arabidopsis ACT DOMAIN REPEAT9 represses glucose signaling pathways. Plant Physiol 192: 1532–1547
- Hsieh WY, Wang HM, Chung YH, Lee KT, Liao HS, Hsieh MH* (2022) THIAMIN REQUIRING2 is involved in thiamin diphosphate biosynthesis and homeostasis. Plant J 111: 1383–1396
- Lee KT, Chung YH, Hsieh MH* (2022) The Arabidopsis glutamine synthetase2 mutants (gln2-1 and gln2-2) do not have abnormal phenotypes. Plant Physiol 189: 1906–1910
- Liao HS, Yang CC, Hsieh MH* (2022) Nitrogen deficiency- and sucrose-induced anthocyanin biosynthesis is modulated by HISTONE DEACETYLASE15 in Arabidopsis. J Exp Bot 73: 3726-3742
- Liao HS, Chung YH, Hsieh MH* (2022) Glutamate: A multifunctional amino acid in plants. Plant Sci 318: 111238
- Hsieh PH, Chung YH, Lee KT, Wang SY, Lu CA, Hsieh MH* (2021) The rice PALE1 homolog is involved in the biosynthesis of vitamin B1. Plant Biotechnol J 19: 218-220
- Liao HS, Chung YH, Chardin C, Hsieh MH* (2020) The lineage and diversity of putative amino acid sensor ACR proteins in plants. Amino Acids 52: 649-666
Domestic
- 2024: National Innovation Award, Institute for Biotechnology and Medicine Industry
- 2009: Career Development Award, Academia Sinica
