[Chuan-Chih Hsu] TIMAHAC: Streamlined Tandem IMAC-HILIC Workflow for Simultaneous and High-Throughput Plant Phosphoproteomics and N-glycoproteomics
POST: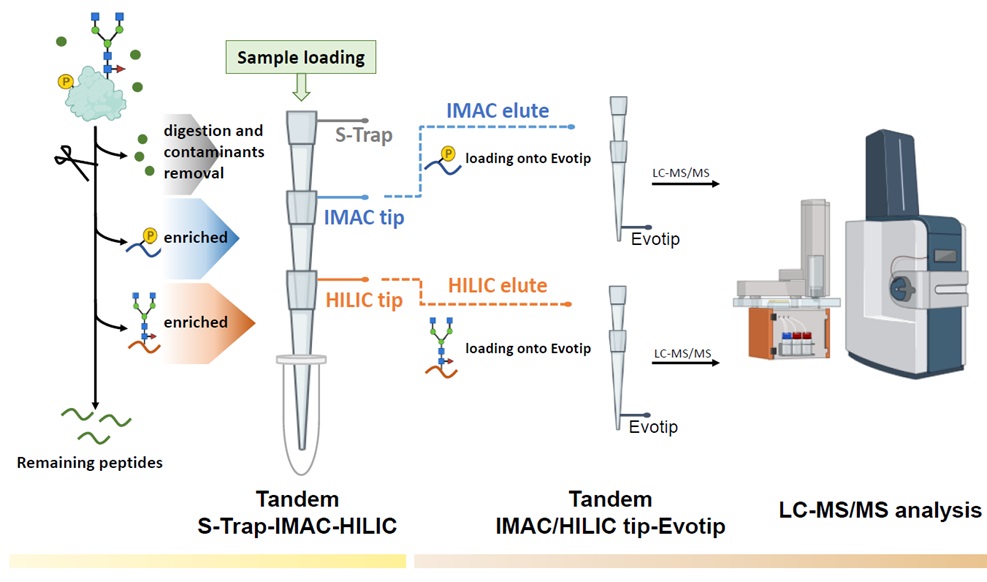
Experimental design of the TIMAHAC workflow for simultaneous analysis of plant phosphoproteomics and N-glycoproteomics. Contaminants removal and protein digestion are conducted within an S-Trap micro column. Phosphopeptides and N-glycopeptides are concurrently enriched through centrifugation using the tandem S-Trap-IMAC-HILIC strategy. The enriched peptides are directly loaded onto an Evotip and subsequently analyzed using an Evosep system coupled with a timsTOF HT mass spectrometer in DDA mode.
Protein post-translational modifications (PTMs) are crucial in plant cellular processes, particularly in protein folding and signal transduction. N-glycosylation and phosphorylation are notably significant PTMs, playing essential roles in regulating plant responses to environmental stimuli. However, current sequential enrichment methods for simultaneous analysis of phosphoproteome and N-glycoproteome are labor-intensive and time-consuming, limits their throughput. Addressing this challenge, the Proteomics Core Lab introduces a novel tandem S-Trap-IMAC-HILIC strategy, termed TIMAHAC, for simultaneous analysis of plant phosphoproteomics and N-glycoproteomics. This approach integrates IMAC and HILIC into a tandem tip format, streamlining the enrichment process of phosphopeptides and N-glycopeptides. The key innovation lies in the use of a unified buffer system and an optimized enrichment sequence to enhance efficiency and reproducibility. The applicability of TIMAHAC was demonstrated by analyzing the Arabidopsis phosphoproteome and N-glycoproteome in response to abscisic acid (ABA) treatment. Up to 1,954 N-glycopeptides and 11,255 phosphopeptides were identified from Arabidopsis, indicating its scalability for plant tissues. Notably, distinct perturbation patterns were observed in the phosphoproteome and N-glycoproteome, suggesting their unique contributions to ABA response. Our results reveal that TIMAHAC offers a comprehensive approach to studying complex regulatory mechanisms and PTM interplay in plant biology, paving the way for in-depth investigations into plant signaling networks.
The first author, Chin-Wen Chen, and the second author, Pei-Yi Lin, are research assistants in the Proteomics Core Lab. The study has been published in the journal of Molecular & Cellular Proteomics.