[Chuan-Chih Hsu] Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics
POST: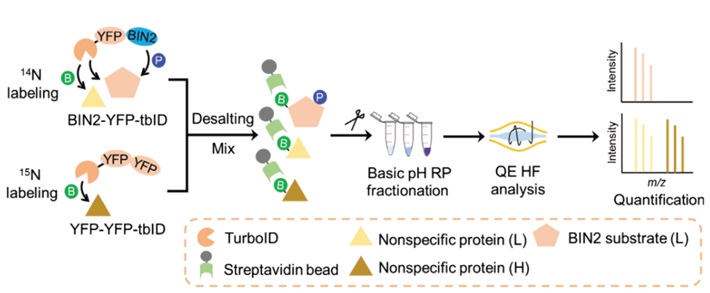
Schematic diagram of the workflow of stable isotope labeling and TurboID-based proximity labeling for the identification of BIN2-proximal proteins in Arabidopsis
Elucidating kinase–substrate relationships in posttranslational modification (PTM) networks is crucial for understanding signal transduction pathways but is technically difficult because kinase–substrate interactions tend to be transient. Here, we demonstrate that TurboID-based proximity labeling (TbPL) effectively and specifically captures the substrates of kinases. TbPL-mass spectrometry (TbPL-MS) identified over 400 proximal proteins of Arabidopsis thaliana BRASSINOSTEROID-INSENSITIVE2 (BIN2). A large portion of the BIN2-proximal proteins showed BIN2-dependent phosphorylation in vivo or in vitro, suggesting that these are candidate BIN2 substrates. Our proteomic analysis establishes the BIN2 signaling network and uncovers BIN2 functions in regulating key cellular processes such as transcription, RNA processing, translation initiation, vesicle trafficking, and cytoskeleton organization. Our work presents a powerful method for mapping PTM networks and important insights into the signaling network that controls key cellular functions underlying plant growth and acclimation.
Kim, Park, and Hsu et al (2023) Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics
Link:https://academic.oup.com/plcell/advance-article/doi/10.1093/plcell/koad013/6993989