[Chuan Ku] Single-cell transcriptomics unveils xylem cell development and evolution
POST:Xylem is the most abundant biological tissue on Earth. It provides mechanical support for trees, transports water to various parts of the plant body, and forms wood, an essential natural resource in our daily life. Therefore, how plants regulate the formation of xylem and how it has evolved over time are two important questions in biology. Anatomy and microscopy can be used to study the morphology and development of xylem cells, but static sections cannot completely capture dynamic developmental processes. Besides, different plants have morphologically similar and distinct xylem cells, but their developmental and evolutionary relationships remain largely unknown.
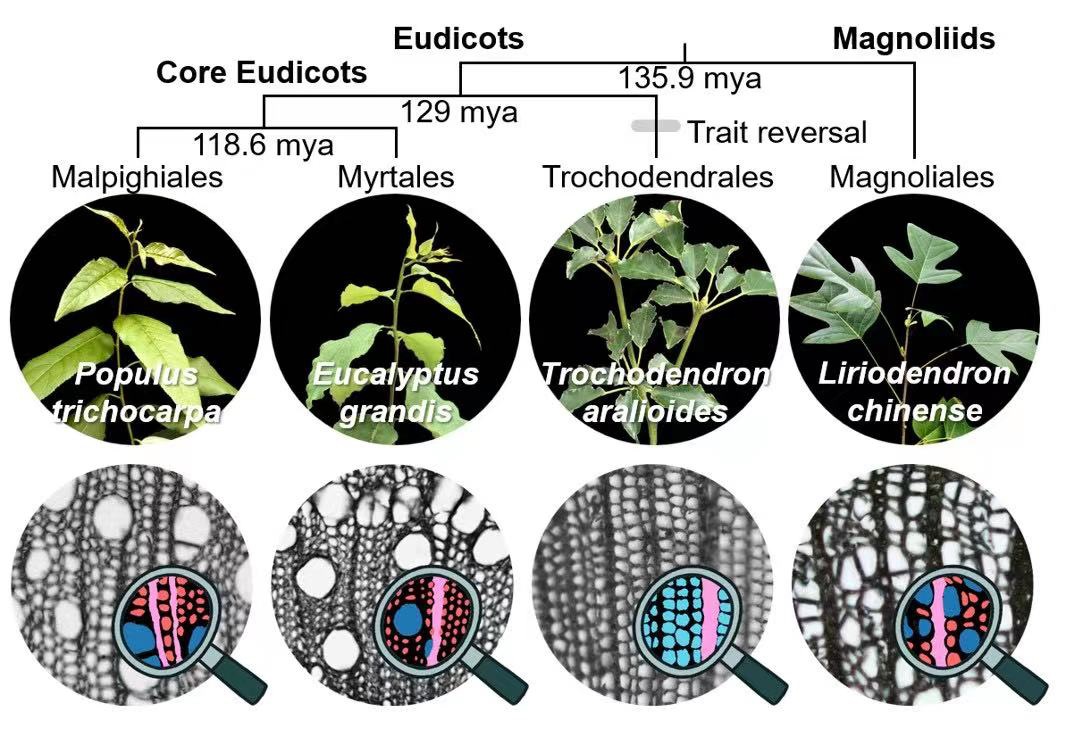
To better understand the formation of xylem, a research team led by Drs. Chuan Ku (Institute of Plant and Microbial Biology, Academia Sinica) and Ying-Chung Jimmy Lin (Department of Life Science and Institute of Plant Biology, National Taiwan University) sampled stem xylem tissue from four tree species and collected transcriptomic data (gene expression profiles) from tens of thousands individual xylem cell protoplasts. Combining single-cell and laser capture microdissection sequencing methods, they were able to map transcriptomes to cells with specific morphology and to reconstruct an atlas of developing xylem cells through clustering and trajectory analyses of single cells. The four species represent different lineages of flowering plant (angiosperm) evolution, including a more basal angiosperm Liriodendron chinense, two core eudicots Populus trichocarpa and Eucalyptus grandis, and a basal eudicot Trochodendron aralioides. The vast majority of angiosperms have libriform fibers for mechanical support, vessel elements for conducting water, and ray parenchyma cells for lateral transport of nutrients. However, T. aralioides, which is native to Taiwan and endemic to East Asia, has undergone an "evolutionary reversal" and does not have libriform fibers and vessel elements, which are replaced by tracheids similar to those seen in gymnosperms. This work shows that the developmental trajectories of ray cells are highly conserved across the four woody plants. Although the libriform fibers and vessel elements of L. chinense are morphologically similar to those in eudicots, they have their distinct developmental lineages. It was also found that the tracheids of T. aralioides exhibit strong transcriptomic similarity to vessel elements rather than libriform fibers of other eudicots. Through cross-species comparisons, this work not only provides important information on xylem formation, but also opens up new perspectives for research on plant tissue evo-devo.
This work has been published in the journal Genome Biology. In addition to the aforementioned ones, the authors’ affiliations include Genome and Systems Biology Degree Program of National Taiwan University and Academia Sinica, National Yang Ming Chiao Tung University, National Chung Hsing University, Taiwan, and Fujian Agriculture and Forestry University, China.
Publication: Chia-Chun Tung, Shang-Che Kuo, Chia-Ling Yang, Jhong-He Yu, Chia-En Huang, Pin-Chien Liou, Ying-Hsuan Sun, Peng Shuai, Jung-Chen Su, Chuan Ku and Ying-Chung Jimmy Lin (2023): Single-cell transcriptomics unveils xylem cell development and evolution. Genome Biology 24:3.
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02845-1