[Pao-Yang Chen] MethylC-analyzer: A comprehensive downstream pipeline for the analysis of genome-wide DNA methylation
POST: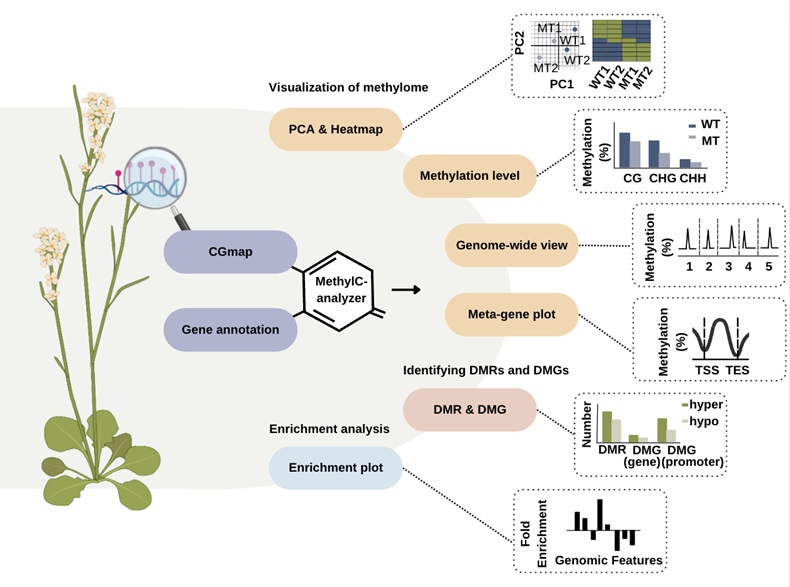
Figure. Schematic diagram of MethyC-analyzer. MethylC-analyzer is a sequential pipeline for analyzing post-align BS-seq and EM-seq.To run MethylC-analyzer, users provide a description text file with post-alignment methylation summaries for each cytosine site of samples, e.g. CGmap and gene annotation file (GTF). The first step MethylC-analyzer is to generate summary methylome figures of CG, CHG, and CHH context, including PCA, heatmap, and the distribution of methylation in each chromosome and gene-centric manner. The following is to perform differential methylation analysis between two groups, for example, identifying differential methylation regions (DMRs) and their related genes (DMGs). Also, comparing methylation status around specific regions. The last step is to visualize the above analysis, MethylC-analyzer will generate publication-ready figures for each step.
DNA methylation is a crucial epigenetic modification involved in multiple biological processes and diseases. Current approaches for measuring genome-wide DNA methylation via bisulfite sequencing (BS-seq) include whole-genome bisulfite sequencing (WGBS), reduced representation bisulfite sequencing (RRBS), and enzymatic methyl-seq (EM-seq). The recent post-alignment methylation tools are specialized for the interpretation of CG methylation, which is known to dominate mammalian genomes, however, non-CG methylation (CHG and CHH, where H refers to A, C, or T) is commonly observed in plants and fungi and is closely associated with gene regulation, transposon silencing, and plant development. Thus, we have developed a MethylC-analyzer to analyze and visualize post-alignment WGBS, RRBS, and EM-seq data focusing on CG. The tool is able to also analyze non-CG sites to enhance deciphering genomes of plants and fungi. By processing aligned data and gene location files, MethylC-analyzer generates a genome-wide view of methylation levels and methylation in user-specified genomic regions. The meta-plot, for example, allows the investigation of DNA methylation within specific genomic elements. Moreover, our tool identifies differentially methylated regions (DMRs) and investigates the enrichment of genomic features associated with variable methylation. MethylC-analyzer functionality is not limited to specific genomes, and we demonstrated its performance on both plant and human data. MethylC-analyzer is a Python- and R-based program designed to perform comprehensive downstream analyses of methylation data, providing an intuitive analysis platform for scientists unfamiliar with DNA methylation analysis. It is available as either a standalone version for command-line uses or as a docker image, publicly accessible at http://paoyang.ipmb.sinica.edu.tw/Software.html#SERVICE
Link: https://as-botanicalstudies.springeropen.com/articles/10.1186/s40529-022-00366-5