[Pao-Yang Chen] ATACgraph: profiling genome wide chromatin accessibility from ATAC-seq
POST: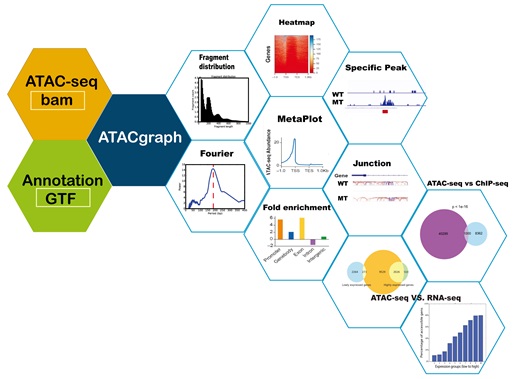
ATACgraph is a software specifically designed for analyzing ATAC-seq. Its functions include filtering out reads from mitochondrial or plastid DNA, outputting the distribution of fragment lengths, and calculating the periodicity of fragment length distribution for quality check, identifying chromatin open regions and genomic features enrichment analysis, comparing two ATAC-seq data, and associating between accessible genes and gene expression, etc.
Assay for transposase-accessible chromatin using sequencing data (ATAC-seq) is an efficient and precise method for revealing chromatin accessibility across the genome. Most of the current ATAC-seq tools follow chromatin immunoprecipitation sequencing (ChIP-seq) strategies that do not consider ATAC-seq-specific properties. To incorporate specific ATAC-seq quality control and the underlying biology of chromatin accessibility, we developed a bioinformatics software named ATACgraph for analyzing and visualizing ATAC-seq data. ATACgraph profiles accessible chromatin regions and provides ATAC-seq specific information including definitions of nucleosome free regions (NFR) and nucleosome occupied regions. ATACgraph also allows identification of differentially accessible regions between two ATAC-seq datasets. ATACgraph incorporates the docker image with the Galaxy platform to provide an intuitive user experience via the graphical interface. Without tedious installation processes on a local machine or cloud, users can analyze data through activated websites using pre-designed workflows or customized pipelines composed of ATACgraph modules. Overall, ATACgraph is an effective tool designed for ATAC-Seq for biologists with minimal bioinformatics knowledge to analyze chromatin accessibility. ATACgraph can be run on any ATAC-seq data with no limit to specific genomes. As validation, we demonstrated ATACgraph on human genome to showcase its functions for ATAC-seq interpretation. This software is publicly accessible and can be downloaded at https://github.com/RitataLU/ATACgraph.