[Chih-Horng Kuo] Species boundaries and molecular markers for the classification of 16SrI phytoplasmas inferred by genome analysis
POST: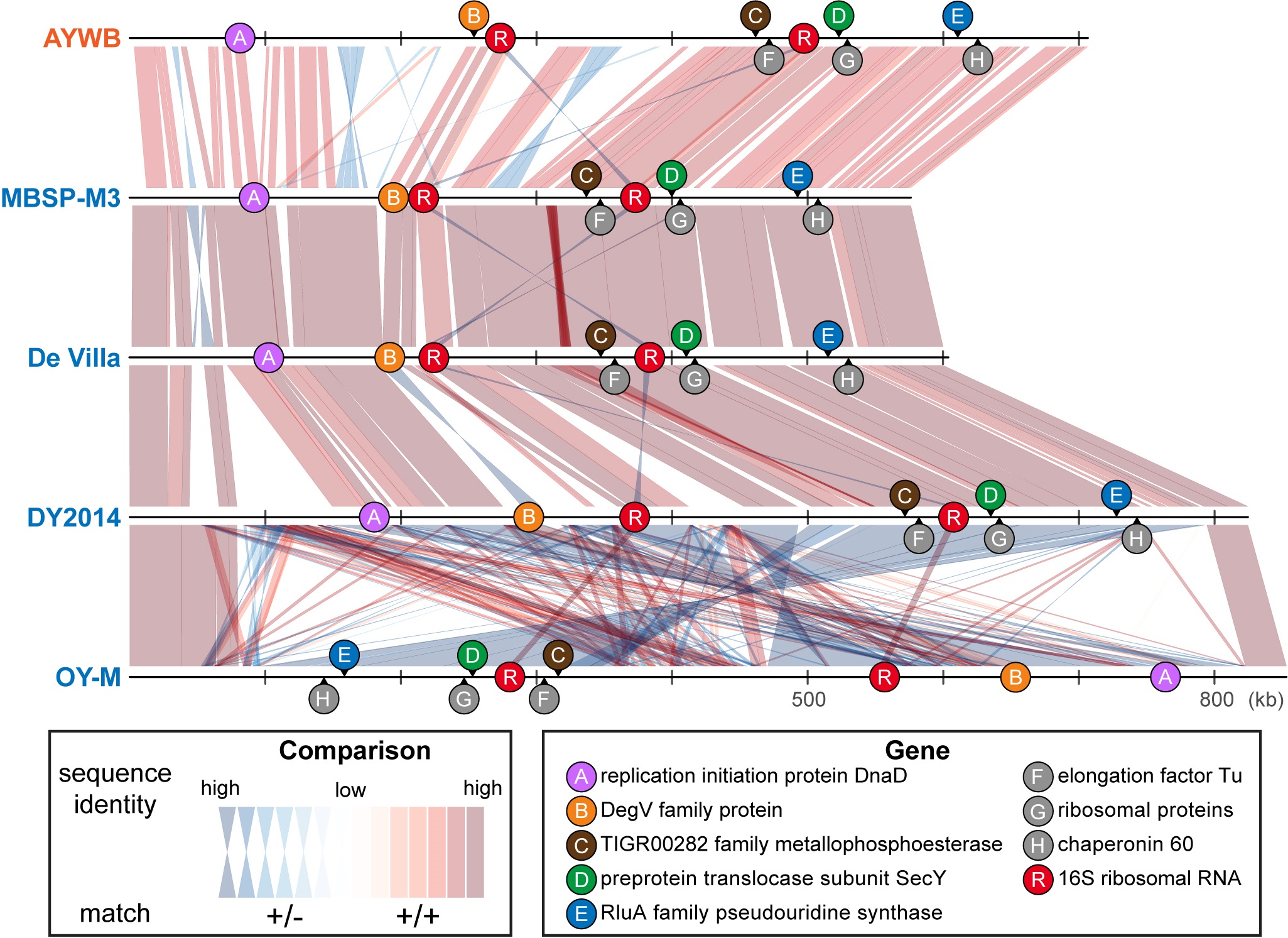
Genome alignment and chromosomal location of marker genes among 16SrI phytoplasmas.
Source: Cho et al. (2020) Front Microbiol. DOI: 10.3389/fmicb.2020.01531
Classification and identification of microbes have always been fundamental topics in biological research. For example, when a new infectious disease occurs, rapid and accurate identification of the pathogen is a key step in formulating effective control strategies. Moreover, when infectious diseases occur in different areas, populations, hosts, or cause different symptoms, confirming whether these diseases are caused by the same or different pathogens is also a critical information for the control and further research. However, for the pathogens that are difficult to isolate or cultivate, such as the phytoplasmas that impact agriculture worldwide, classification and identification are difficult. For these plant-pathogenic bacteria that have remained uncultivated despite decades of efforts, the commonly adopted classification system is the one that was developed in the 1990s and based on the restriction fragment length polymorphism (RFLP) analysis of bacterial 16S rRNA genes. Although this system is easy to use, it has several severe shortcomings. For example, one strain may harbor two 16S rRNA genes that are classified into different taxonomic groups, or different strains may harbor highly similar 16S rRNA genes while diverge in other genes in their genomes. As such, improvements in the accuracy and resolution of phytoplasma identification and classification are important.
With the recent advancements in DNA sequencing and genomics, whole genome sequencing is becoming feasible even for uncultivated bacteria. Due to their impacts on agriculture, the genome sequences of >10 strains belonging to the 16SrI group of phytoplasmas have been determined. With this valuable data set, this project conducted whole-genome comparisons to better understand these bacteria. The results demonstrated that although these bacteria were classified as one single species and all share >99% sequence identity in their 16S rRNA genes, the other genes that they have in their genomes and the sequence identity of their shared genes are highly variable. As such, these strains could be reclassified into three different species. Within the same species, strains share >78% of their genes, and the whole genome average nucleotide identities (ANI) are >97%. For cross-species comparisons, strains share <78% of their genes and have <94% ANI. These findings are important to basic research such as bacteria taxonomy and applied research such as plant pathology.
However, for practical purposes, whole genome sequencing of bacteria remains costly and time-consuming, particularly for uncultivated ones. In order to contribute to applications, this project also examined all of the shared genes in these phytoplasmas, and selected five genes with high discriminatory power for distinguishing these different species to develop PCR primers as new molecular markers. All of these five new markers could be sequenced in one single Sanger sequencing reaction, and are distributed evenly across the chromosome. Therefore, these new markers provide a cost-effective system that could greatly improve the accuracy of classification and identification of field samples.
This work is done in collaboration between Dr. Chih-Horng Kuo and Dr. Saskia Hogenhout (John Innes Centre, UK). The first author Ms. Shu-Ting Cho is a Project Manager in the Kuo lab. The manuscript was published as a part of a special topic “Mollicutes: From Evolution To Pathogenesis” (https://www.frontiersin.org/research-topics/9908/mollicutes-from-evolution-to-pathogenesis) hosted by Frontiers in Microbiology.
Cho ST, Kung HJ, Huang W, Hogenhout SA, Kuo CH* (2020) Species boundaries and molecular markers for the classification of 16SrI phytoplasmas inferred by genome analysis. Frontiers in Microbiology 11: 1531. DOI: 10.3389/fmicb.2020.01531