[Chuan Ku] Potential and challenges of single-cell RNA sequencing for microbial eukaryotes
POST:
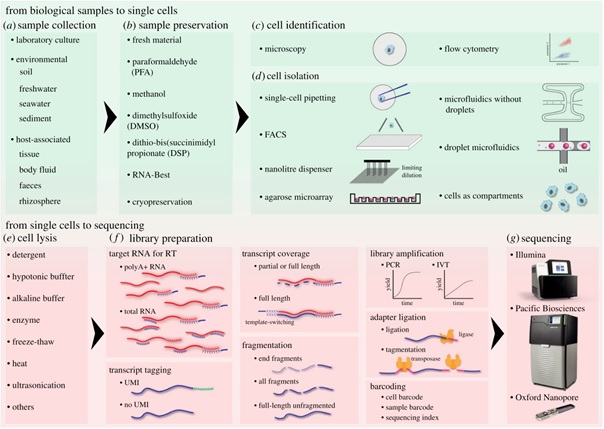
Single-cell RNA sequencing enables quantification and characterization of gene transcripts in individual cells — the basic structural and functional unit of life. Recent advances in its methodology and efficiency have revolutionized our understanding of cell types and developmental processes in multicellular organisms, particularly vertebrates. As shown in a few studies, the same approach can also be applied to microbial eukaryotes, or protists, which encompass the vast majority of eukaryotic diversity and are integral components of various ecosystems. In this article, Chuan Ku at IPMB and Arnau Sebé-Pedós at the Centre for Genomic Regulation (CRG) in Barcelona reviewed the current methods of single-cell RNA sequencing and discussed its potential and challenges in studies on microbial eukaryotes. Single-cell RNA sequencing is divided into key steps to provide a quick overview for researchers interested in this technology, with special focus on possible technical difficulties for diverse unicellular eukaryotes. With newly available sequencing platforms and analysis tools, they further offered future perspectives on how transcriptomic information of isolated single cells can facilitate studies on cellular states and microbial interactions in both simple lab cultures and complex field communities.
This article is part of the theme issue “Single cell ecology” published on October 7 in Philosophical Transactions of the Royal Society B: https://royalsocietypublishing.org/doi/10.1098/rstb.2019.0098