[Matzke, Antonius; Matzke, Marjori; Chen, Pao-Yang] Global impacts of chromosomal imbalance on gene expression in Arabidopsis and other taxa
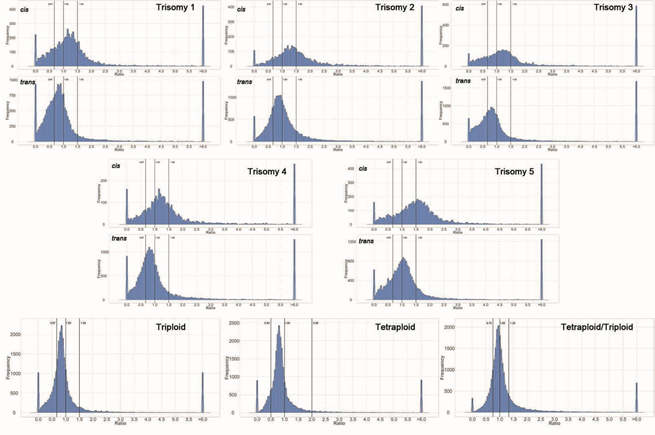
POST:The sequencing reads from RNA-Seq were averaged for the biological replicates. For each expressed gene, a ratio of the averaged read counts in the respective experimental (trisomic or ploidy) genotype was made over the read counts in the diploid control. These ratios were plotted in bins of 0.05. The X axis notes the value for each bin and the Y axis notes the number of genes per bin. For the five trisomics, genes were partitioned into those encoded on the varied chromosome (cis) versus those encoded on the remainder of the genome that was not varied in dosage (trans). A ratio of 1.00 represents no change in the experimental genotype versus the diploid. A ratio of 1.50 represents a gene dosage effect in cis whereas 1.00 represents dosage compensation. A ratio of 0.67 represents the inverse ratio of gene expression in trans. These ratio values are demarcated with labeled vertical lines. The triploid and tetraploid ploidy series were analyzed in the same manner for all expressed genes. The vertical demarcations in this case correspond to the respective direct or inverse relationship of the ploidy comparison. The tetraploid/triploid comparison was generated by producing the respective ratios and plotting the distribution. Each comparison is labeled in the respective panel.
Changes in dosage of part of the genome (aneuploidy) have long been known to produce more severe phenotypic consequences than changes in the number of whole genomes (ploidy). To study the basis of these differences, we examined, global gene expression in mature leaf tissue for all five trisomics and in diploids, triploids and tetraploids of Arabidopsis thaliana. The trisomics produced a greater spread of expression modulation than the ploidy series. In general, expression of genes on the varied chromosome ranged from compensation to dosage effect whereas genes from the remainder of the genome ranged from no effect to reduced expression approaching the inverse level of chromosomal imbalance (2/3). Genome-wide DNA methylation was investigated in each genotype and found to shift most prominently with trisomy 4 but otherwise exhibited little change, indicating that genetic imbalance is usually mechanistically unrelated to DNA methylation. Independent inspection of gene functional classes demonstrated that ribosomal, proteasomal and gene body methylated genes were less modulated compared to all classes of genes whereas genes encoding transcription factors, signal transduction components and organelle-targeted proteins were more tightly inversely affected. Comparing transcription factors and their targets in the trisomics and in expression networks revealed considerable discordance, illustrating that altered regulatory stoichiometry is a major contributor to genetic imbalance. Re-analysis of published data on gene expression in disomic yeast and trisomic mouse cells detected similar stoichiometric effects across broad phylogenetic taxa and indicated that these responses reflect normal gene regulatory processes.