
Teh, Ooi-Kock (鄭惠國)
Assistant Research Fellow
- Ph.D., Plant Sciences, University of Oxford, UK
- Bryophytes, cell biology, developmental biology, Physcomitrium patens
- okteh@gate.sinica.edu.tw
- okteh@as.edu.tw
- +886-2-2787-1102 (Lab: )
- +886-2-2787-1101 (Office: )
- Lab Website
- ORCID
- Web of Science (WOS)
- Google Scholar
We use combinatory approaches of genetics, cell biology, biochemistry to understand what makes the plants look the way they are.
For crustaceans such as crabs and lobsters, their hard armoured protective shells restrict their body growth and shedding this layer of exoskeleton through moulting is a critical phase in their lives. Plants face a similar problem during their growth as their cells are encased in stiff cell walls which are rich in polysaccharides (which are long chains of carbohydrate molecules) such as cellulose and pectins. So how do plant cells achieve cellular growth while being trapped inside the walls? Lacking the ability to move, plants cannot just wiggle themselves out from the walls before expansion like the crustaceans do. On the contrary, plants have acquired unique mechanisms that enable them to remodel their cell walls to soften the wall for cellular expansion. Before we could begin to understand this mechanism, it is important to take a look at the cell wall structures:
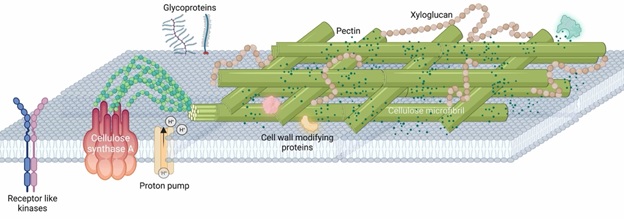
The plant cell wall lies on the outside of the cell and is a complex matrix of polysaccharides that are consist of cellulose microfibril, pectins and hemicellulose (e.g. xyloglucan). These polysaccharides are the predominant compositions and contribute to 95% of the molecular weight. The remaining 5% are proteins that are highly decorated with sugar moieties - known as glycoproteins - which play elusive signalling roles. In close proximity to the cell wall is plasma membrane which is fluidic and dynamic. Many protein complexes that are necessary to coordinate plant growth and cell wall remodelling lie on the plasma membrane. For example, receptor-like kinases could sense changes in the wall integrity and relay such information to the cell so that the cell could respond to it. Proton pumps on the other hand help to create an acidic environment in the wall which is crucial for the wall loosening.
The main compositions of plant cells walls are cellulose microfibril, hemicellulose and pectins which provide the fundamental support as the plant exoskeleton. Despite its significance as a major global carbon sink (where final photosynthetic products are stored), plant biologists still cannot agree on the arrangement of these carbon-rich molecules in the wall. Scattered throughout the walls are some proteins which are highly decorated with sugars, collectively called glycoproteins. These glycoproteins play mysterious roles in plant growth as overexpressing them often leads to peculiar changes in the plant growth. There are also receptor proteins kinases which are embedded within the plasma membrane but span beyond the cell wall-plasma membrane interface to help relay signalling information from the wall to the inside of the cell. Some famous receptor proteins such as FERONIA and WAK (wall associated kinases) are known to interact with polysaccharides from the cell wall and signal important changes downstream to alert the plant cells to cope with the ever-changing environment.
When associating the cell wall structures with cellular growth in plants, a few questions become imminently apparent:
(1) When plant cells are ready to expand, how do the cells instruct the cell walls to loosen up? Subsequently, how is the loosening halted when cell expansion has ceased?
(2) As the cell wall is being remodelled (softening/rigidification) to accommodate for cellular growth, how are compositional changes within the wall sensed by the cell?
Figuring out these answers is not easy and requires robust tools as well as a model than can be genetically manipulated. Our lab is primarily using Physcomitrium patens (P. patens), a moss as a model to answer the above questions. Moss belongs to the bryophyte group, which is a sister group to all the vascular plants on land.
Most plant biologists agree that land plants evolved from the fresh water algae in the Cambrian era (515-494 million years ago). Splitting from a common ancestor, the bryophytes represent the earliest plants that conquered and survived on the barren land. Imagine arriving at a highly desiccated environment compared to its previous “water world”, the bryophytes must have invented some tricks that allows them to survive on the dry land. These tricks are likely to be cell wall-related and aim to prevent unregulated water loss. Hence, moss is a valuable system for the investigation of cell wall signalling, especially during the early phase of land plant evolution.
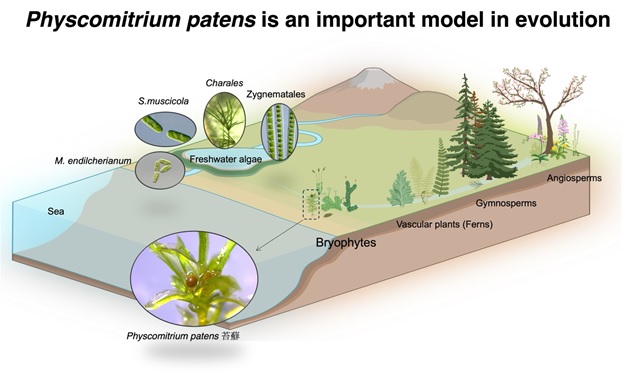
Land plants are thought to have evolved from a class of unicellular and filamentous fresh water algae called Zygnematales. Extant bryophytes (which are non-vascular plants) split from the vascular plants during the Cambrian era (515-494 million years ago) and form a monophyletic sister group that consists of 3 main groups: mosses, liverworts and hornworts. The commonly used moss model, Physcomitrium patens was sequenced in 2008 (Rensing et al., 2008) and can be genetically manipulated using various tools that have been developed.
Using the moss, we have recently made some progress:
(1) We showed that a cell wall glycoprotein called SLEEPING BEAUTY (SB) could affect the signalling output of an auxin-dependent transcription factor, named ARFC (Teh et al., 2022). The ARFC would control the expression of cell wall modifying enzymes such as PECTIN METHYLESTERASES (PMEs). The PMEs are ubiquitous cell wall enzymes that modify and determine the degree of methylation in the cell wall pectins. Pectins with varying degree of methylation are often used to explain the stiffness of the cell wall and hence its methylation status is a crucial regulator during cell wall remodelling. We have begun to investigate the functions of PME in moss and found that overexpression of particular PMEs could have a profound effect on the plant growth (unpublished), suggesting that pectin dynamics a key driver for increased architectural complexity.
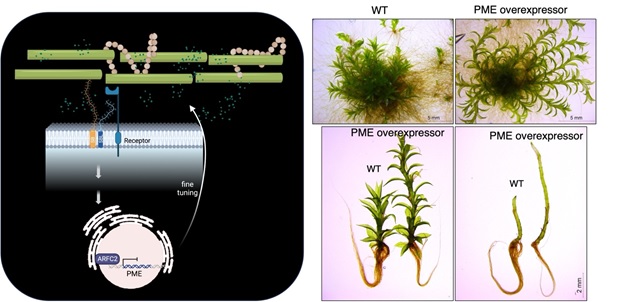
Left panel: A schematic diagram that summarises the function of SLEEPING BEAUTY in regulating cell wall remodelling. Right panel: Moss transgenic plants that overexpress a specific copy of PME showed enhanced growth in the leaf-like gametophores compared to wild-type (WT) control.
(2) We recently demonstrated that a type of small signalling peptide, Rapid Alkalinisation Factor (RALF) is a key factor for apical tip growth in the filamentous structures of moss (Febrianto and Teh et al., 2022). Moss colonies that are defective of RALFs could not elongate properly and gave rise to colonies with smooth edges. Intriguingly, the RALF peptide exhibited a polarized localiation at the growing tip. As RALFs in the flowering plant Arabidopsis are known to bind to FERONIA, a well-known cell wall integrity sensor, we are pursuing the connection between RALFs and cell wall integrity regulation.
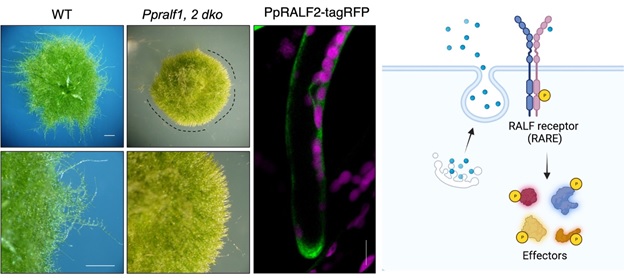
Left panel: A moss mutant Ppralf1,2 dko that is defective of Rapid Alkalinisation Factor (RALF) exhibited a smooth growing edge (indicated by the dotted lines) as compared to wild-type control. This indicates that tip growth is compromised in the Ppralf1,2 dko mutant and that RALF is necessary to promote tip growth in moss. In support of this, the one of these RALFs, PpRALF2 (as indicated by the green signal) is found to be localised to the tip of a growing filamentous protonemata. Magenta signals in the image indicate chloroplasts in the cells. Right panel: A schematic representation that summarises the action of RALFs (blue spheres) which are secreted to the outside of the cell. The RALFs are expected to be perceived by a cell surface-localised receptor RARE (RALF receptor) which became phosphorylated and goes on to relay the information to downstream effectors by phosphorylating them.
The first step in our investigation is to identify the FERONIA-equivalent in the moss. We have taken an explorative phosphoproteomic approach using mass spectrometry to analyse peptides that became phosphorylated when RALF is overexpressed. Receptor-like kinases that became phosphorylated are termed RAREs (RALF receptors). We identified 13 RAREs and intriguingly one of these RAREs, RARE13, when overexpressed in tobacco leaves leads to cell death (unpublished). We are currently investigating how a growth-promoting kinase could cause an opposite phenotype when present in a large quantity.
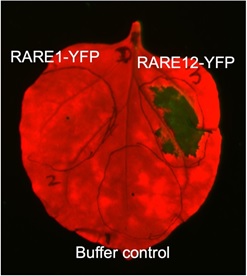
A tobacco leaf is infiltrated with Agrobacterium suspensions that contain either RARE1 or RARE12 that are fused with a yellow fluorescent protein (YFP) reporter. Note that only areas that are infiltrated with RARE12 displayed cell lesions (or cell death, as indicated by the dark green signal), while areas infiltrated with RARE1-YFP and buffer only control appeared unaffected.
All publications
Selected Publications
- Ooi, S-E., Sarpan, N., Taranenko, E, Feshah, I., Nuraziyan, A., Roowi, S.H., Burhan, M.N., Jayanthi, N., Rahmah, A.R.S., Teh, O-K., Ong-Abdullah, M., Tatarinova, T.V. (2023). Small RNAs and Karma methylation in Elaeis guineensis mother palms are linked to high clonal mantling. Plant Molecular Biology, 111: 345-363. http://doi.org/10.1007/s11103-022-01330-4
- Teh, O-K., Singh, P., Ren, J., Huang, L-T., Ariyarathne, M., Salamon, B.P., Wang, Y., Kotake, T., Fujita, T. (2022). Surface-localized glycoproteins act through class C ARFs to fine-tune gametophore initiation in Physcomitrium patens. Development 149, dev200370. https://doi.org/10.1242/dev.200370
- Ginanjar, E.F. , Teh, O-K., Fujita, T. (2022) Characterisation of rapid alkalinisation factors (RALFs) in Physcomitrium patens reveals functional conservation in tip growth. New Phytologist, 233 (6):2442-2457. https://doi.org/10.1111/nph.17942
- Bao L, Inoue N, Ishikawa M, Gotoh E, Teh O-K, Higa T, Morimoto T, Ginanjar EF, Harashima H, Noda N, Watahiki M, Hiwatashi Y, Sekine M, Hasebe M, Wada M, Fujita T. (2022) A PSTAIRE-type cyclin-dependent kinase controls light responses in land plants. Science Advances. 28:8(4):eabk2116. https://doi.org/1126/sciadv.abk2116
- Brillada C*, Teh OK*, Ditengou FA, Lee CW, Klecker T, Saeed B, Furlan G, Zietz M, Hause G, Eschen-Lippold L, Hoehenwarter W, Lee J, Ott T, Trujillo M. (2021) Exocyst subunit Exo70B2 is linked to immune signaling and autophagy. Plant Cell. Apr 17;33(2):404-419. doi: 10.1093/plcell/koaa022. *Co-first authorship
- Do TH, Pongthai P, Ariyarathne M, Teh OK, Fujita T. (2020) AP2/ERF transcription factors regulate salt-induced chloroplast division in the moss Physcomitrella patens. J Plant Res. Jul;133(4):537-548. doi: 10.1007/s10265-020-01195-y.
- Teh OK, Lee CW, Ditengou FA, , Klecker T, Furlan G, Zietz M, Hause G, Eschen-Lippold L, Hoehenwarter W, Lee J, Ott T, Trujillo M. (2019) Phosphorylation of the exocyst subunit Exo70B2 contributes to the regulation of its function. bioRxiv doi: https://doi.org/10.1101/266171.
- Shimada T, Fuji K, Ichino T, Teh OK, Koumoto Y, Hara-Nishimura I. (2018) GREEN FLUORESCENT SEED, to Evaluate Vacuolar Trafficking in Arabidopsis Seeds. Methods Mol Biol. 1789:1-7. doi: 10.1007/978-1-4939-7856-4_1.
- Liu H, Ravichandran S, Teh OK, McVey S, Lilley C, Teresinski HJ, Gonzalez-Ferrer C, Mullen RT, Hofius D, Prithiviraj B, Stone SL. (2017) The RING-Type E3 Ligase XBAT35.2 Is Involved in Cell Death Induction and Pathogen Response. Plant Physiol. Nov;175(3):1469-1483. doi: 10.1104/pp.17.01071.
- Teh OK, Hatsugai N, Tamura K, Fuji K, Tabata R, Yamaguchi K, Shingenobu S, Yamada M, Hasebe M, Sawa S, Shimada T, Hara-Nishimura I. (2015) BEACH-domain proteins act together in a cascade to mediate vacuolar protein trafficking and disease resistance in Arabidopsis. Mol Plant. Mar;8(3):389-98. doi: 10.1016/j.molp.2014.11.015.
- Munch D*, Teh OK*, Malinovsky FG*, Liu Q, Vetukuri RR, El Kasmi F, Brodersen P, Hara-Nishimura I, Dangl JL, Petersen M, Mundy J, Hofius D. (2015) Retromer contributes to immunity-associated cell death in Arabidopsis. Plant Cell. Feb;27(2):463-79. doi: 10.1105/tpc.114.132043. *Co-first authorship
- Teh OK, Hofius D. (2014) Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J Exp Bot. Mar;65(5):1297-312. doi: 10.1093/jxb/ert441.
- Teh OK, Shimono Y, Shirakawa M, Fukao Y, Tamura K, Shimada T, Hara-Nishimura I. (2013) The AP-1 μ adaptin is required for KNOLLE localization at the cell plate to mediate cytokinesis in Arabidopsis. Plant Cell Physiol. Jun;54(6):838-47. doi: 10.1093/pcp/pct048.
- Teh OK, Ramli US. (2011) Characterization of a KCS-like KASII from Jessenia bataua that elongates saturated and monounsaturated stearic acids in Arabidopsis thaliana. Mol Biotechnol. Jun;48(2):97-108. doi: 10.1007/s12033-010-9350-x.
- Teh OK, Moore I. (2007) An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature. Jul 26;448(7152):493-6. doi: 10.1038/nature06023.
Domestic
- 2024: Career Development Award , Academia Sinica

