[Chih-Horng Kuo/Erh-Min Lai/Chih-Hang Wu] Comparative transcriptomics reveals context- and strain-specific regulatory programs of Agrobacterium during plant colonization
POST:Agrobacterium is widely known for its ability to transfer DNA into plant genomes, a process that forms the basis of Agrobacterium-mediated transformation (AMT), an essential tool in plant biotechnology. Most studies, however, have focused on a few model strains under laboratory conditions, leaving open questions about how diverse natural strains behave inside plants.
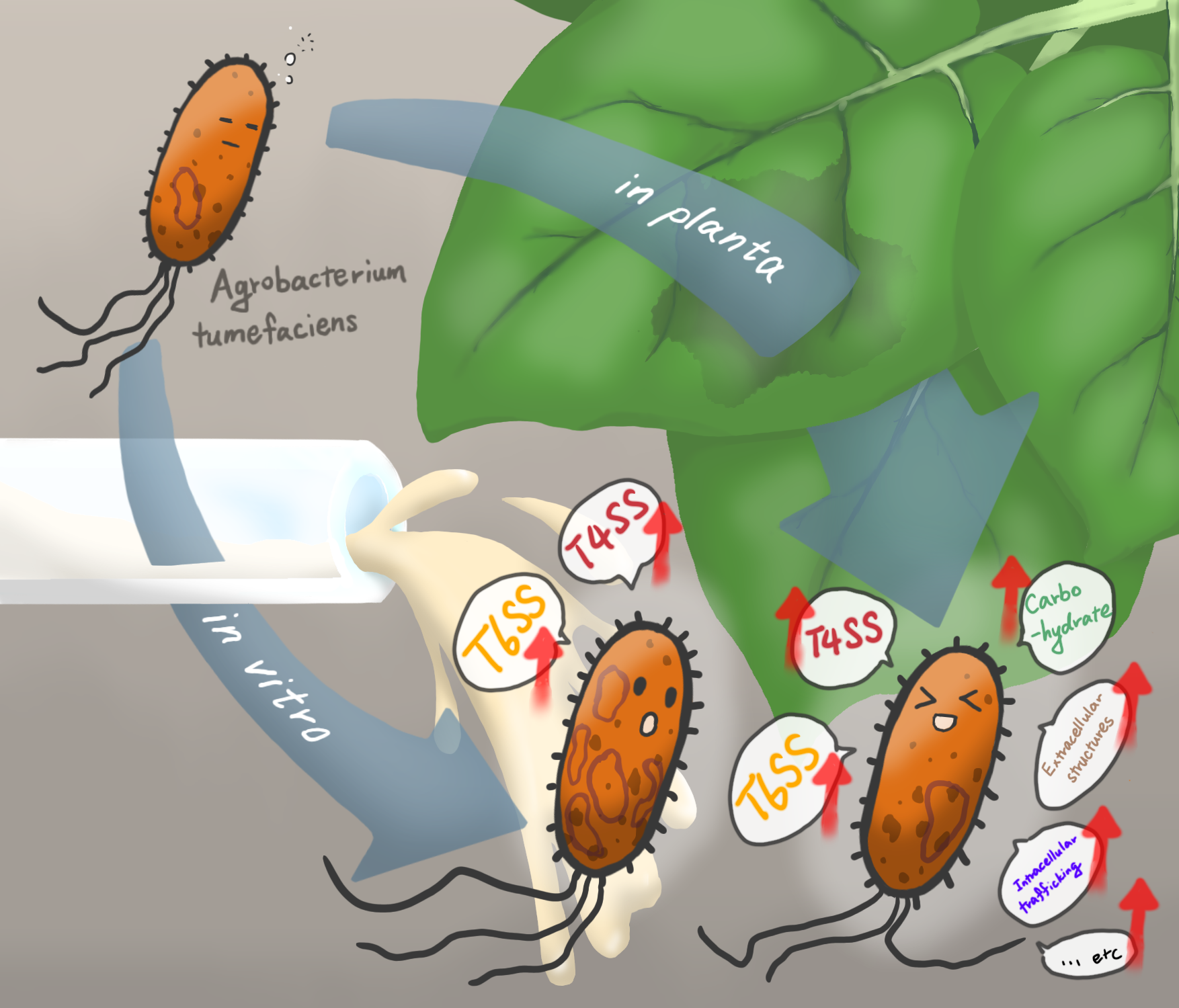
In this new study, researchers investigated the gene expression programs of several wild-type Agrobacterium strains and identified strain 1D1108 as particularly effective in both stable transformation in legumes and transient transformation in Nicotiana benthamiana. Using RNA-Seq, the team examined the transcriptome of 1D1108 in different environments, focusing on those related to virulence induction, including acidic pH, the chemical inducer acetosyringone, and inside the leaf tissue of Nicotiana benthamiana.
The results revealed that the plant environment triggered far more extensive gene regulation than laboratory treatments alone, with over a thousand genes specifically regulated. These included genes for bacterial attachment, secretion systems, and nutrient uptake, highlighting a complex and integrated response to host cues. Comparisons with other Agrobacterium strains showed little overlap in gene expression, underscoring how regulatory programs are both context-dependent and highly strain-specific.
To broaden the perspective, the team also compared Agrobacterium with Pseudomonas syringae, another plant pathogen. Both activated their specialized secretion systems inside plants, but their overall gene expression patterns were distinct, reflecting different ecological strategies.
Together, these findings demonstrate that studying Agrobacterium in real plant contexts and incorporating diverse strains is essential to capture its full biology and improve transformation efficiency. This approach not only overcomes the limitations of artificial conditions and model strains but also opens new opportunities for future applications.
This study is a collaboration among the teams of Drs. Chih-Hong Kuo, Erh-Min Lai, and Chih-Hang Wu in the Institute of Plant and Microbial Biology, Academia Sinica. The first author Dr. Yu Wu was a doctoral student in the Taiwan International Graduate Program on Molecular and Biological Agricultural Science (TIGP-MBAS). The co-author Ms. Hsing-Yi Chang is a research assistant supervised by Dr. Kuo. This project was supported by Academia Sinica Grand Challenge Program and the National Science and Technology Council of Taiwan.