[Chih-Horng Kuo] Comparative genome analysis of 'Candidatus Phytoplasma luffae' reveals the influential roles of potential mobile units in phytoplasma evolution
POST: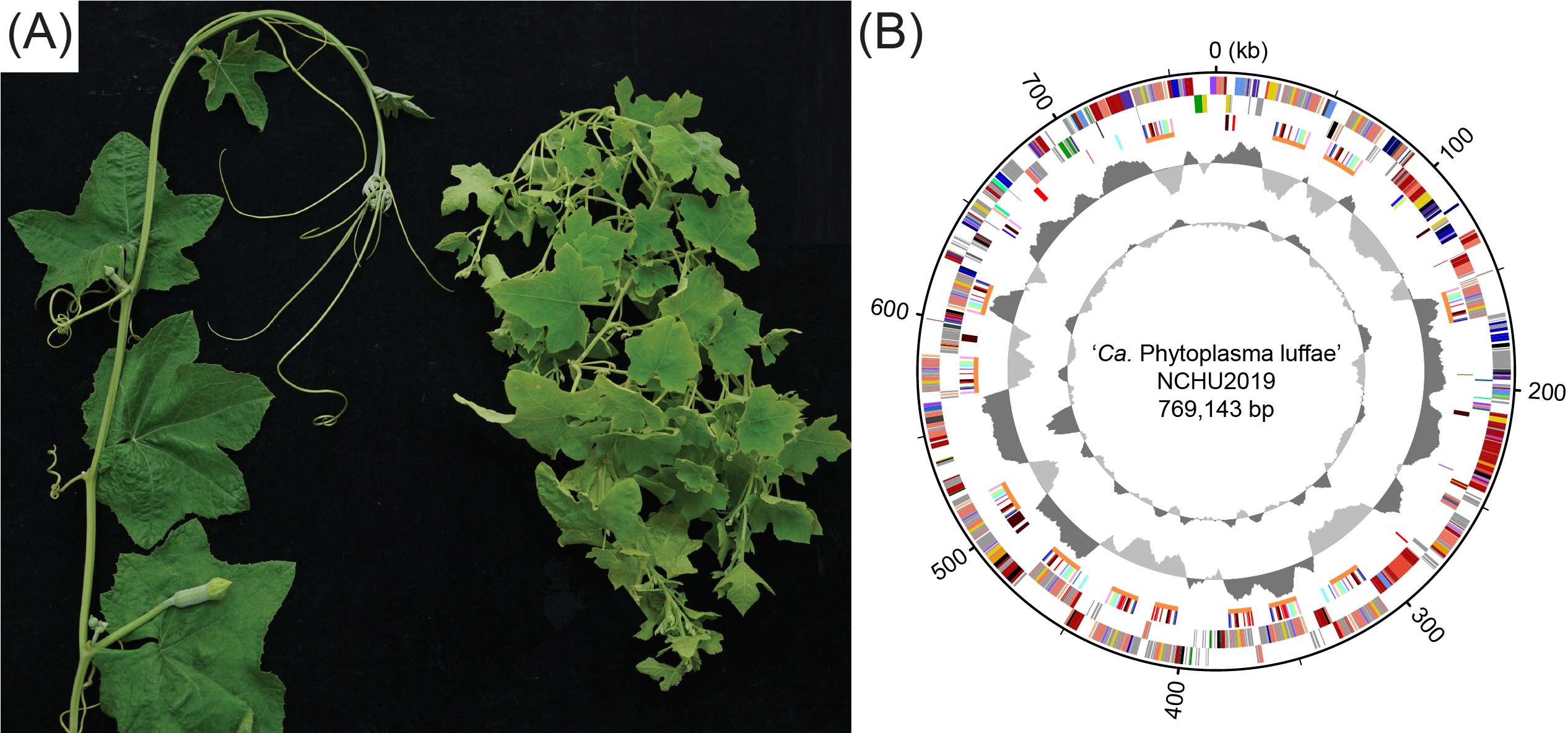
Figure. Witches’ broom disease of loofah. (A) Infection symptoms. Left, healthy control; right, phytoplasma-infected. (B) Genome map of ‘Candidatus Phytoplasma luffae’ strain NCHU2019 found in the plant shown in panel A.
Phytoplasmas are plant-pathogenic bacteria that are distributed widely and cause substantial agricultural losses in the world. In Taiwan, the crops that are impacted by phytoplasma-associated diseases include peanut, loofah, periwinkle, pear, papaya, and sweet potato. Due to the infeasibility of establishing axenic culture of phytoplasmas, it is difficult to study these bacteria using conventional microbiology tools. To overcome this limitation, Drs. Chih-Horng Kuo (Institute of Plant and Microbial Biology, Academia Sinica, Taiwan) and Jun-Yi Yang (Institute of Biochemistry, National Chung Hsing University, Taiwan) collaborated to investigate the evolution and pathogenicity mechanisms using genomics and molecular biology tools.
In this newly published study, the team utilized an infected plant collected in Central Taiwan as the material for conducting whole-genome sequencing and analysis of a ‘Canadidatus Phytoplasma luffae’ strain associated with loofah witches' broom disease. The genomic characterization identified at least 13 potential mobile units (PMUs), which is a new high record among the phytoplasma genome sequences characterized to date. Because the effector genes involved in phytoplasma pathogenicity are often associated with PMUs, the proliferation of these mobile genetic elements also increased the number of effector genes in the genome, which may promote evolutionary processes that lead to subfunctionalization or neofunctionalization of these virulence factors. Furthermore, based on comparative analysis conducted at the genus level, the PMUs among different phytoplasmas can be classified into three major types based on gene organization. Notably, PMU abundance explains nearly 80% of the variance in phytoplasma genome sizes. These findings not only established a foundation for future research on PMUs, but also provided the first quantitative estimate for the importance of PMUs in phytoplasma genome variability.
This research article was published in Frontiers in Microbiology, the first author Ching-Ting Huang and co-authors Shu-Ting Cho and Yu-Chen Lin are research assistants supervised by Chih-Horng Kuo. The funding was provided by the Ministry of Science and Technology, the Ministry of Education, and Academia Sinica in Taiwan.
Huang CT, Cho ST, Lin YC, Tan CM, Chiu YC, Yang JY*, Kuo CH* (2022) Comparative genome analysis of 'Candidatus Phytoplasma luffae' reveals the influential roles of potential mobile units in phytoplasma evolution. Frontiers in Microbiology 13:773608. DOI: 10.3389/fmicb.2022.773608 [bioRxiv preprint DOI: 10.1101/2021.09.09.459700]