[Chih-Horng Kuo] Modular evolution of secretion systems and virulence plasmids in a bacterial species complex
POST: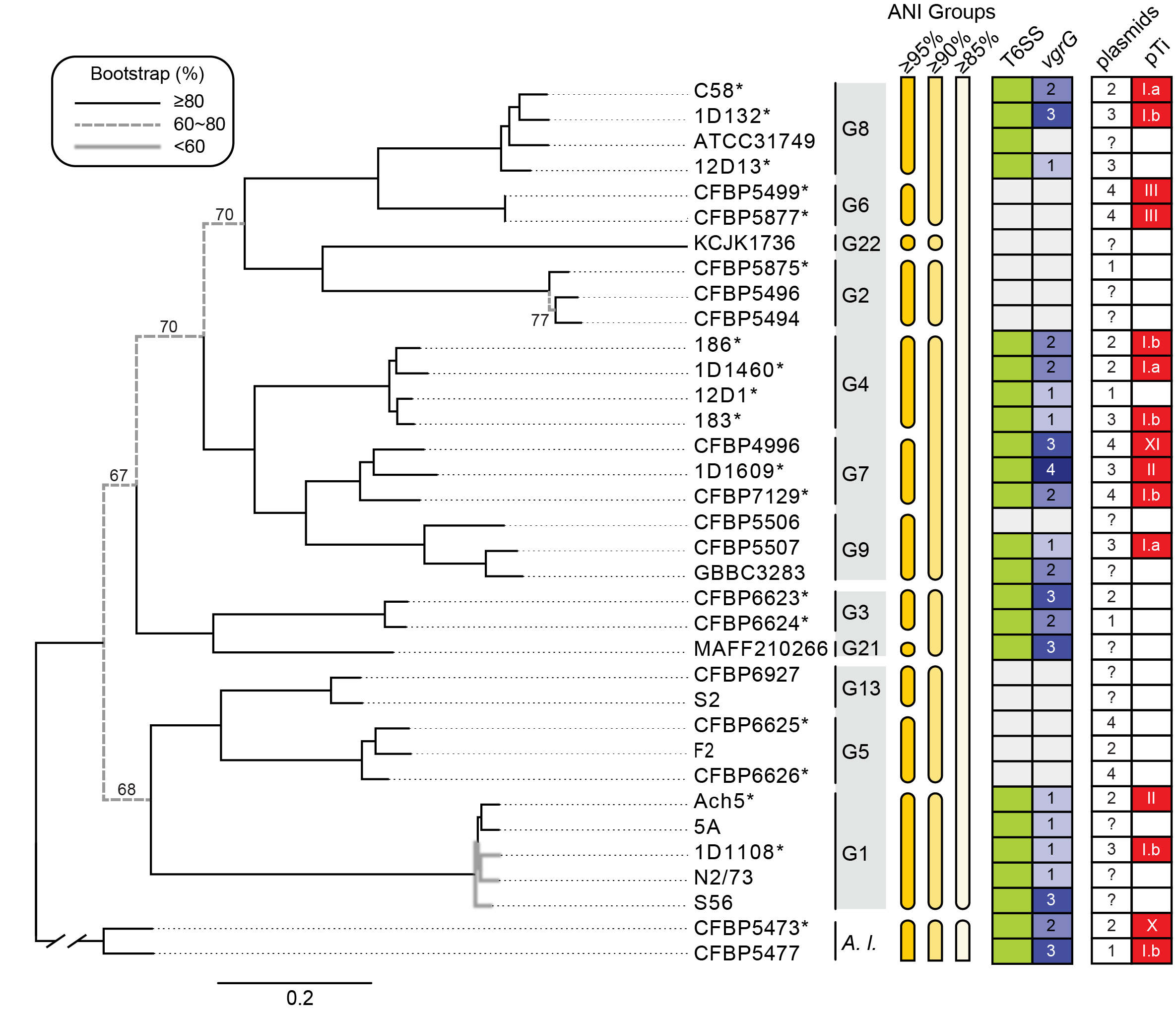
Figure legend. Relationships among representatives of the Agrobacterium tumefaciens species complex. The genomospecies assignments (i.e., G1-G9, G13, and G21-G22), grouping based on genome-wide average nucleotide identity (ANI), presence/absence of type VI secretion system (T6SS)-encoding gene cluster (green: present; white: absent), copy number of vgrG (white background: absent), number of plasmids, and the tumor-inducing plasmid (pTi) type (white background: absent) are labeled.
In biology, “species” is the basic unit for description and research of different organisms. However, many named species as defined in current bacterial taxonomy correspond to species complexes, in which the species boundaries and relationships among species are uncertain. Such uncertainties not only hamper research, but can also impact the identification of pathogens or even policy-making for quarantine regulations. To solve this challenge, an international team led by Chih-Horng Kuo in the Institute of Plant and Microbial Biology (Academia Sinica, Taiwan) utilized the Agrobacterium tumefaciens species complex as the study system and devised strategies for genomic analysis. These bacteria are known for their phytopathogenicity, as well as a critical tool for genetic manipulation in plant sciences and agricultural biotechnology.
The results indicated that genomic analysis can clearly delineate the species boundaries within a species complex based on DNA sequences or gene content, which allows for the investigation of genetic differentiation at the among- and within-species levels. For analysis related to microbial ecology, the type VI secretion system that is utilized by Agrobacterium for inter-bacterial competition was found to have experienced a complex history of gene gains and losses, and these evolutionary processes have shaped the diversity of effectors and related genes. For the tumor-inducing plasmids and the type IV secretion system that are required for Agrobacterium phytopathogenicity, the genomic analysis also revealed a complex evolutionary history and a high level of genetic diversity, which may explain the differences in virulence against various plant hosts among these strains. In summary, the findings can facilitate future functional studies of Agrobacterium, and the newly developed strategies for multi-level genomic analysis are applicable to other bacterial species complexes.
This research article was published in BMC Biology. The two co-first authors, Lin Chou and Yu-Chen Lin, are both research assistants supervised by Chih-Horng Kuo. The collaborating teams include those led by Erh-Min Lai (Academia Sinica, Taiwan) and Jeff Chang (Oregon State University, USA). The funding was supported by Academia Sinica (Taiwan), Ministry of Science and Technology (Taiwan), and Department of Agriculture (USA).
Chou L#, Lin YC#, Haryono M, Santos MNM, Cho ST, Weisberg AJ, Wu CF, Chang JH, Lai EM, Kuo CH* (2022) Modular evolution of secretion systems and virulence plasmids in a bacterial species complex. BMC Biology 20:16. #Equal contribution.