[Chuan Ku] Insights into genome evolution of giant viruses
POST:Giant viruses of eukaryotes are a diverse group of viruses known for their gigantic genomes. Using large-scale network and comparative analyses of gene sharing, the research team of Dr. Chuan Ku at Institute of Plant and Microbial Biology found that the patterns of giant virus genome variation often contradict with the phylogeny of a few core genes and instead highly correlate with the diversity and types of eukaryotic hosts. This discovery improves our understanding of the role of hosts in viral genome gigantism and sheds light on further studies on the origin(s) of giant viruses and their coevolution with eukaryotes. The work has been published in the journal Virus Evolution.
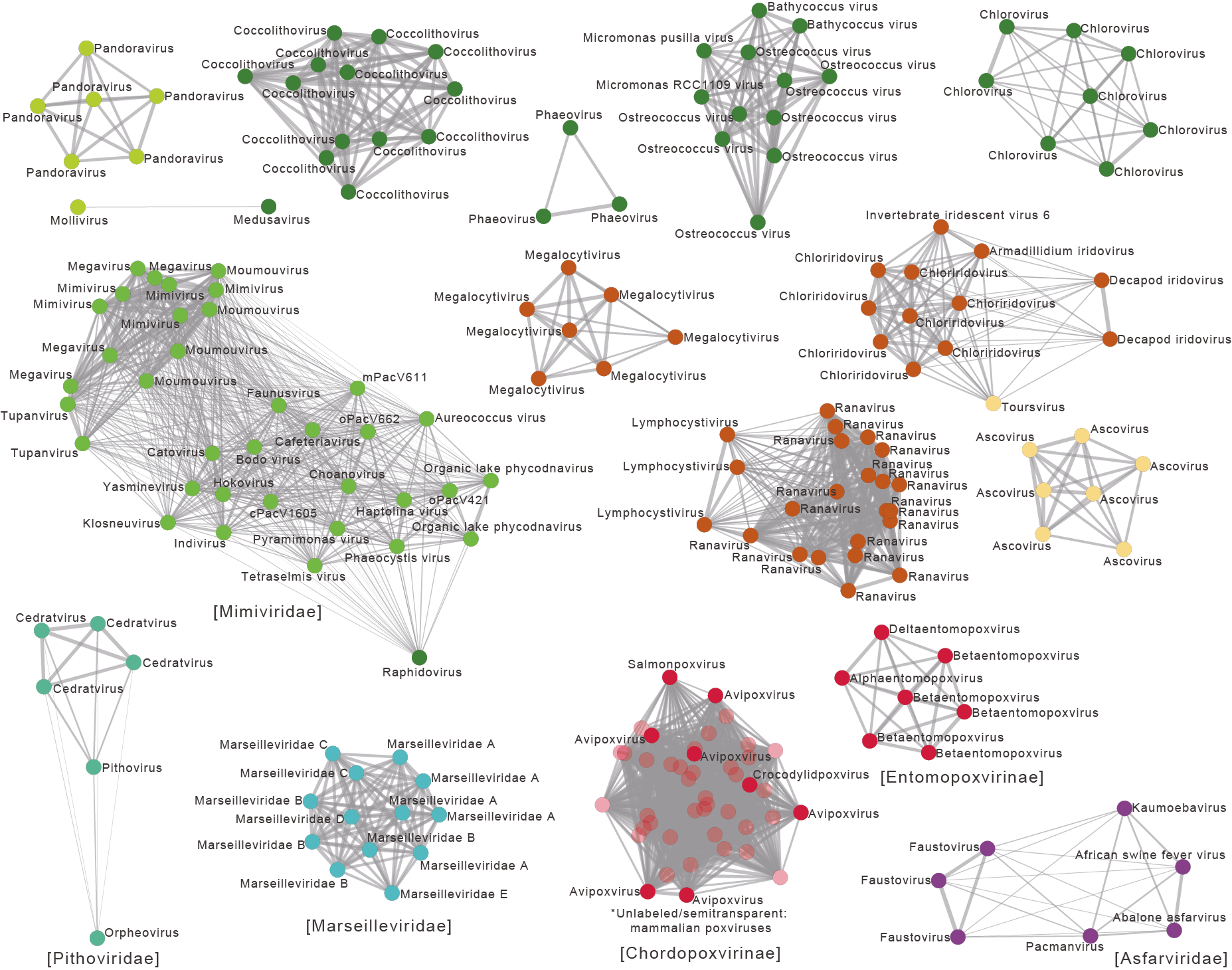
Viruses are parasitic particles that can only replicate through host cells. Most viruses, such as SARS-CoV-2, carry a few to tens of genes in their genomes, but viruses in the newly established phylum Nucleocytoviricota can encode hundreds to over two thousand genes in their double-stranded DNA genomes, outnumbering those of many bacteria. Commonly called giant viruses, members of Nucleocytoviricota have been isolated from across major groups of eukaryotes, including many animals, algae, amoebae and flagellates. How and why giant virus genomes have become so gigantic remains a major outstanding question in evolutionary biology.
Phylogenetic trees based on a few widely distributed core genes have been commonly used to understand the evolutionary relationships among giant viruses and form the basis of delineating higher-level taxa such as classes, orders and families. Through whole-genome analyses of homologous genes between viruses, this study reveals networks of individual genomes and family pangenomes with patterns of gene sharing that contradict with the core-gene tree topology, in particular at higher taxonomic levels. For example, the two families in the class Pokkesviricetes, Asfarviridae and Poxviridae, have lower gene sharing between them than each with some other families. The study further shows that the host diversity of different families correlates with their intrafamilial gene repertoire variation and that viruses that are from different families but infect a similar host type (algae, vertebrates or insects) tend to share more genes, such as those encoding ion channels or proteins involved in animal immune responses. By exerting selection or providing a pool of genes that can be transferred, hosts, their environment and their associated microbes shape the gene contents of giant viruses and are potentially a crucial factor in viral genome gigantism and the origin(s) of giant viruses. With the ever-increasing number of giant viruses discovered in various environments and their recognition as important ecological players, the comprehensive approach in this study will further better our understanding of giant virus genomes and evolution.
The first author of this work is Tsu-Wang Sun, who is currently a graduate student in Genome and Systems Biology Degree Program, Academia Sinica and National Taiwan University, and received Undergraduate Research Grant from the Ministry of Science and Technology (MOST) to work on this project. Dr. Chuan Ku is also supported by a MOST research grant and the Academia Sinica Career Development Award.
Publication: Tsu-Wang Sun and Chuan Ku (2021): Unraveling gene content variation across eukaryotic giant viruses based on network analyses and host associations. Virus Evolution 7: veab081.
Website: https://doi.org/10.1093/ve/veab081