[Matzke, Antonius; Matzke, Marjori] A collection of pre-mRNA splicing mutants in Arabidopsis thaliana
POST: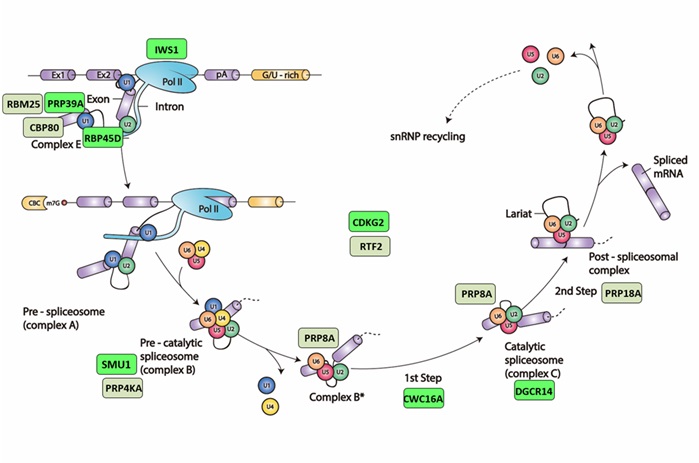
Figure: Spliceosomal cycle and factors identified in the forward genetic screen Splicing is catalyzed by the spliceosome, a large and dynamic ribonucleoprotein (RNP) machine located in the nucleus. Spliceosomes comprise five small nuclear (sn) RNPs, each containing a heptameric ring of Sm or Like-Sm proteins and a different snRNA (U1, U2, U4, U5 or U6), as well as numerous other non-snRNP proteins. During the splicesomal reaction cycle, the five snRNPs act sequentially on the pre-mRNA with a changing assemblage of non-snRNP proteins to form a series of complexes that catalyze two consecutive trans-esterification reactions. In complex E, U1 and U2 snRNPs first recognize the 5’ and 3’ splice sites branch points of introns and interact to form pre-spliceosomal complex A. The subsequent addition of preformed U4/U5/U6 tri-snRNP creates pre-catalytic complex B. Ensuing reorganization steps induce release of U1 and U4 snRNPs and conversion of complex B to complex B*, which catalyzes the first reaction yielding the free 5’ exon and lariat 3’-exon intermediates. The newly formed C complex catalyzes the second reaction to achieve intron lariat excision and exon ligation. Lastly, dismantling of the spliceosome frees individual components to assemble anew at the next intron. The positions of factors identified in our screen, as predicted by their orthologs in yeast or metazoans, are indicated by colored rectangles (green, Hyper-GFP; dull green, GFP-weak). Adapted by permission from Springer Nature, Nat. Rev. Mol. Cell Biol. 15: 108–121, A day in the life of the spliceosome, A. G. Matera and Z. Wang, 2014.
To investigate factors influencing pre-mRNA splicing in plants, we conducted a forward genetic screen using an alternatively-spliced GFP reporter gene in Arabidopsis thaliana. This effort generated a collection of sixteen mutants impaired in various splicing-related proteins, many of which had not been recovered in any prior genetic screen or implicated in splicing in plants. The factors are predicted to act at different steps of the spliceosomal cycle, snRNP biogenesis pathway, transcription, and mRNA transport. We have described eleven of the mutants in recent publications. Here we present the final five mutants, which are defective, respectively, in RNA-BINDING PROTEIN 45D (rbp45d), DIGEORGE SYNDROME CRITICAL REGION 14 (dgcr14), CYCLIN-DEPENDENT KINASE G2 (cdkg2), INTERACTS WITH SPT6 (iws1) and CAP BINDING PROTEIN 80 (cbp80). We provide RNA-sequencing data and analyses of differential gene expression and alternative splicing patterns for the cbp80 mutant and for several previously published mutants, including smfa and new alleles of cwc16a, for which such information was not yet available. Sequencing of small RNAs from the cbp80 mutant highlighted the necessity of wild-type CBP80 for processing of microRNA (miRNA) precursors into mature miRNAs. Redundancy tests of paralogs encoding several of the splicing factors revealed their functional non-equivalence in the GFP reporter gene system. We discuss the cumulative findings and their implications for the regulation of pre-mRNA splicing efficiency and alternative splicing in plants. The mutant collection provides a unique resource for further studies on a coherent set of splicing factors and their roles in gene expression, alternative splicing and plant development.