[Pao-Yang Chen] Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi
POST: Ming-Ren YenSecondary metabolites (SM) produced by fungi and bacteria have long been of exceptional interest owing to their unique biomedical ramifications. The traditional discovery of new natural products that was mainly driven by bioactivity screening has now experienced a fresh new approach in the form of genome mining. Several bioinformatics tools have been continuously developed to detect potential biosynthetic gene clusters (BGCs) that are responsible for the production of SM. Although the principles underlying the computation of these tools have been discussed, the biological background is left underrated and ambiguous. In this review, we emphasize the biological hypotheses in BGC formation driven from the observations across genomes in bacteria and fungi, and provide a comprehensive list of updated algorithms/tools exclusively for BGC detection. Our review points to a direction that the biological hypotheses should be systematically incorporated into the BGC prediction and assist the prioritization of candidate BGC.
The first author Phuong Nguyen Tran was the summer intern of TIGP international internship program (TIGP-IIP). This article is co-authored with Dr. Hsiao-Ching Lin from the Institute of Biological Chemistry in Academia Sinica and published in Applied Microbiology and Biotechnology
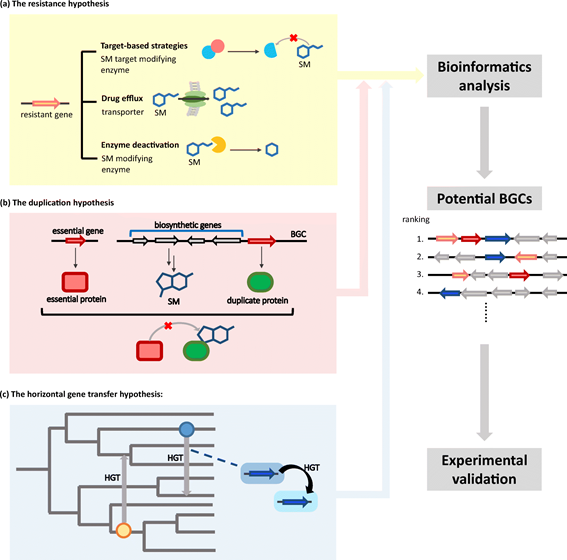
Figure 1: Overview of biological aspects underlying biosynthetic gene cluster (BGC) target-directed detection. Three hypotheses include (a) The resistance hypothesis comprises three notable models: target-based strategies, drug efflux, and enzyme deactivation. (b) The duplication hypothesis holds that the SM producer harbors a protein isoform (duplicate protein) of an essential protein. (c) The horizontal gene transfer hypothesis of core genes is a potential way for microorganism to gain genetic advantage for self-protection.