[Guang-Yuh Jauh] Mutation of Arabidopsis SAURs impairs the efficient translation of transcripts essential for pollen tube growth
POST:
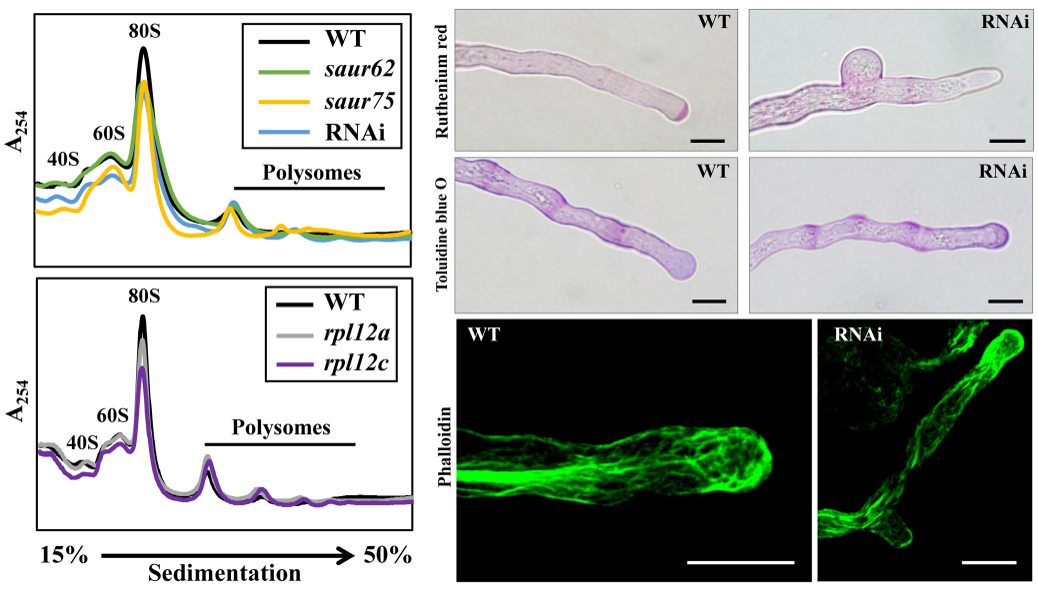
Successful pollen tube elongation is critical for double fertilization. Understanding the biological functions of pollen tube genes will provide new insights into the regulatory machinery underlying this crucial process. He et al. from Dr. Guang-Yuh Jauh’s laboratory identified two Arabidopsis Small Auxin Up RNA genes, SAUR62 and SAUR75, with expression upregulated by pollination according to previous translatomic study (Lin et al. Plant Cell 2014). Both SAUR62 and SAUR75 mainly localized in pollen tube nuclei, and siliques of homozygous saur62 (saur62/-), saur75 (saur75/-), and the SAUR62/75 RNAi knockdown line had many aborted seeds. Pollen viability of these mutants and RNAi lines was normal, but in vitro and in vivo pollen tube growth was defective, with branching phenotypes. Immunoprecipitation with transgenic SAUR62/75-GFP flowers revealed ribosomal protein RPL12 family members as their potential partners, and their individual interactions were further confirmed by yeast two-hybrid (Y2H) and bimolecular fluorescence complementation assay (BiFC). Polysome profiling showed reduced 80S ribosome abundance in homozygous saur62, saur75, rpl12c and SAUR62/75 RNAi flowers, which suggests roles for SAUR62/75 in ribosome assembly. To clarify the roles in translation, total proteins of wild-type (WT) and RNAi flowers were analyzed by iTRAQ, which revealed significantly reduced expression of factors participating in pollen tube wall biogenesis and F-actin dynamics, critical for the elastic properties of tube elongation. Indeed, RNAi pollen tubes showed dislocalization of de-esterified and esterified pectins and F-actin organization. Here, we demonstrate the biological roles of SAUR62/75 and their partners, RPL12 family members, as critical in ribosome assembly for efficient pollen tube elongation and the following fertilization (He et al. Plant Physiology 2018).