[Chin-Min Ho, Shu-Hsing Wu] Serial Spatial Transcriptomes Reveal Regulatory Transitions in Maize Leaf Development.
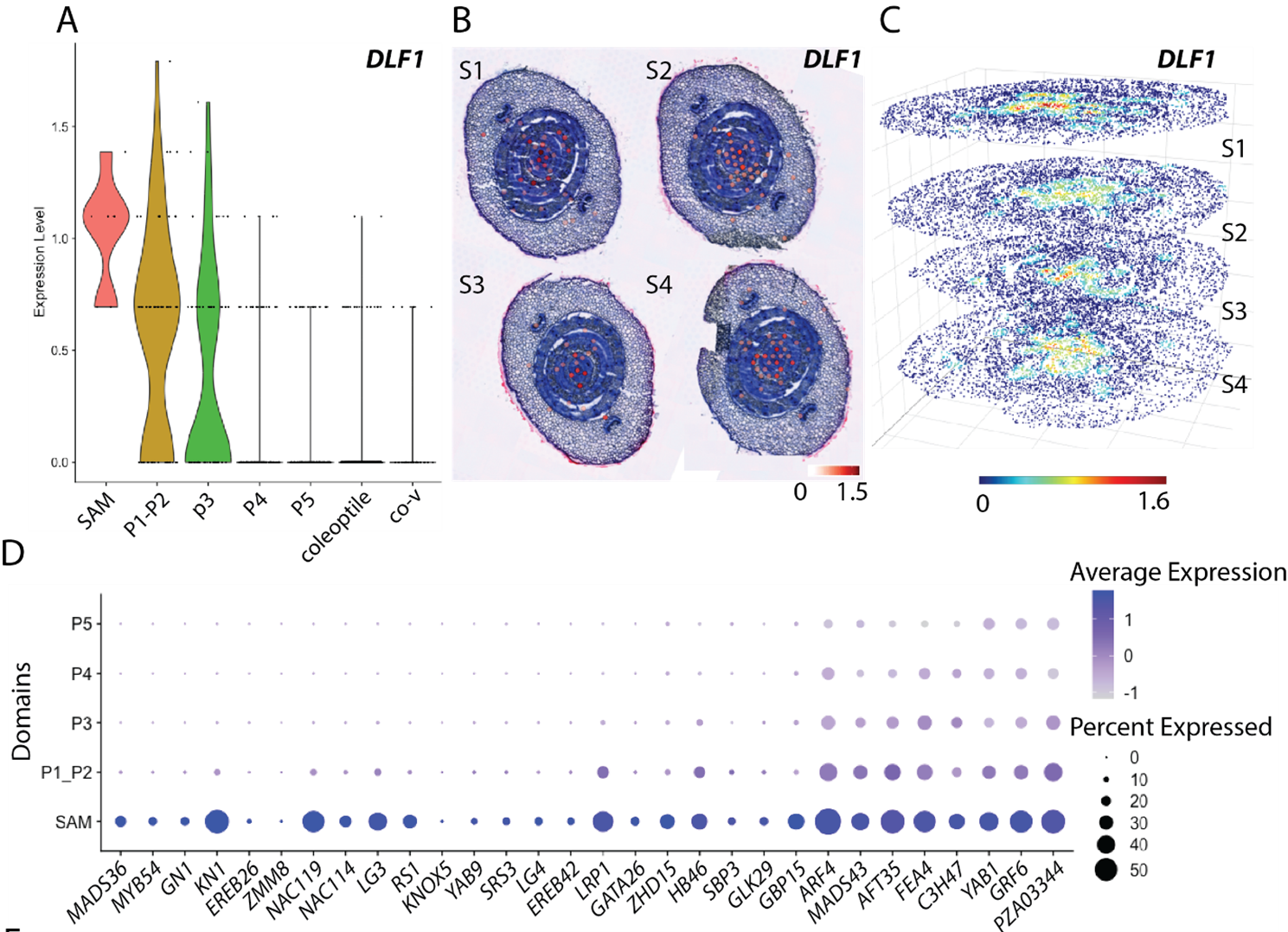
POST:Leaves differentiate progressively from the shoot apical meristem (SAM), yet conventional transcriptomic approaches often lack spatial context and a continuous developmental series, making it difficult to reconstruct the full regulatory progression from the SAM to early leaf primordia and onward to vascular maturation. In this study, we profiled the SAM and sequentially developing young leaves of maize seedlings, optimized the 10× Genomics Visium spatial transcriptomics workflow, and developed a computational toolkit to build three-dimensional (3D) gene expression atlases. By linking cellular states across serial sections, we reconstructed developmental trajectories and captured dynamic transitions from undifferentiated stem-cell populations to functional leaf tissues.
Through integrated spatial–temporal analyses, we identified key regulatory genes associated with meristem maintenance, primordium initiation, vascular differentiation, and tissue heterogeneity. Combined with 3D reconstruction and interactive visualization, our framework provides an explorable spatial expression map and an accompanying analysis pipeline.
We further performed functional validation of candidate transcription factors by generating growth-regulating factor (GRF) mutants in Setaria viridis using CRISPR/Cas9, supporting a regulatory role of GRFs in meristem function and early leaf development.
This work was carried out through a collaborative effort between the Institute of Plant and Microbial Biology and the Biodiversity Research Center, Academia Sinica. The first author, Dr. Chi-Chih Wu, completed the major part of this study during his appointments at these two institutes.