[Ting-Ying Wu] How HSFA1 and Chromatin Dynamics Turn On Plants’ Heat Survival System?
POST: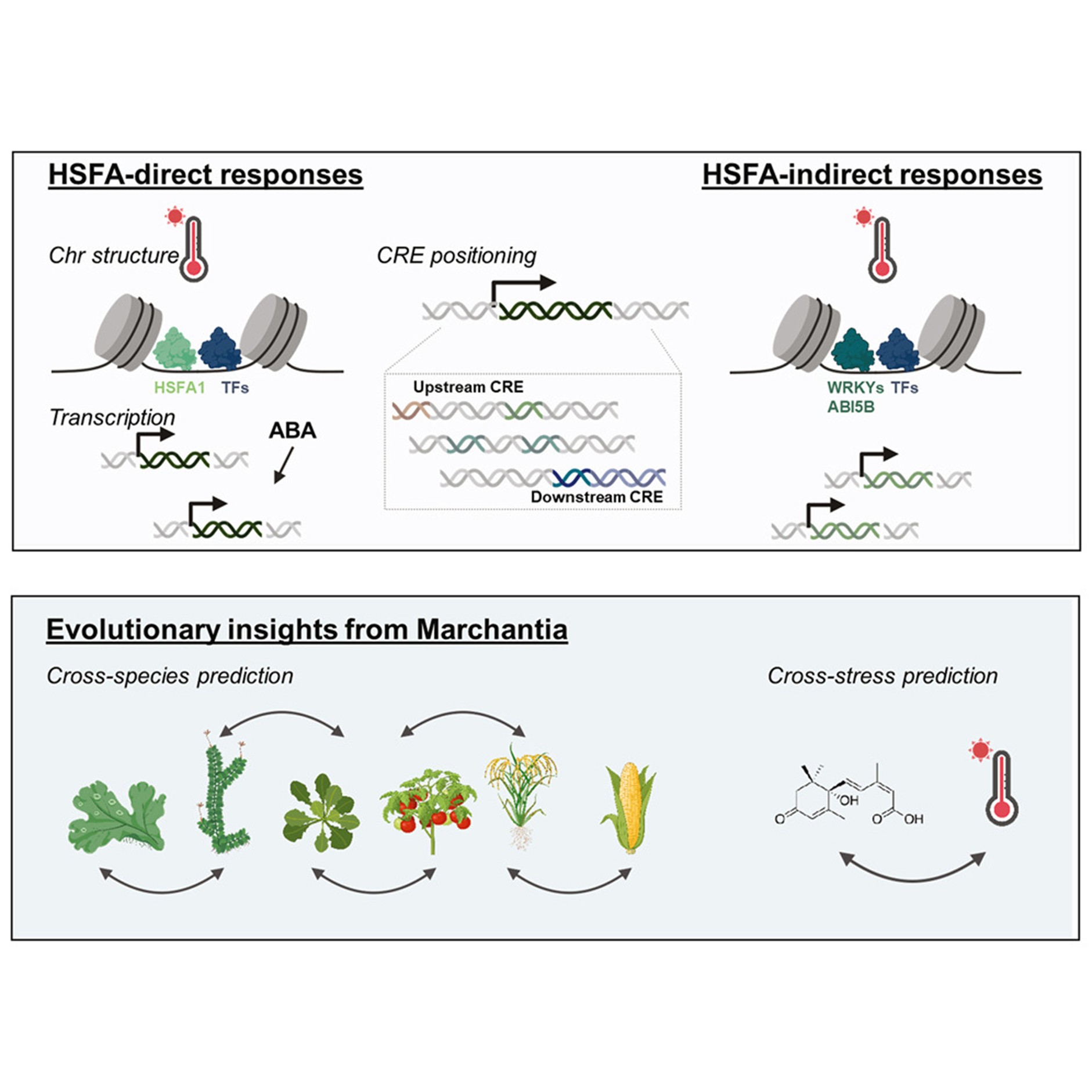
The graphic summarizes the main findings of this study. We found that (I) HSFA1 is required for HS-induced chromatin remodeling, while ABA did not induce genome-wide chromatin changes. (II) MpABI5B and MpWRKY10 are required for MpHSFA1-parallel HS regulation. (III) HSFA1 is required for the accessibility of HS-related CREs in Marchantia. Additionally, we demonstrated that, by using information from CREs, it is possible to predict gene expression at the transcriptional level across plant species and stress types.
Plants, like all living organisms, must respond quickly when their environment becomes stressful, such as during extreme heat. One way they do this is by changing how their DNA is packaged, which helps turn important genes on or off. In this study, we found that a transcription factor called HSFA1 plays a central role in helping plants respond to heat stress. Using both a simple land plant and a flowering plant, we showed that HSFA1 controls which parts of the DNA become accessible during heat stress. This mechanism is highly conserved and is also found in animals, including humans. We also discovered that some plant hormones, such as ABA, influence heat-responsive genes without changing DNA structure. By combining these data with machine learning, we were able to predict how genes respond to stress across species. Our findings reveal a shared and ancient strategy that living organisms use to survive in hot environments.