[Wan-Sheng Lo and Long-Chi Wang] Dimerization of the ETO1 family proteins plays a crucial role in regulating ethylene biosynthesis in Arabidopsis thaliana
POST: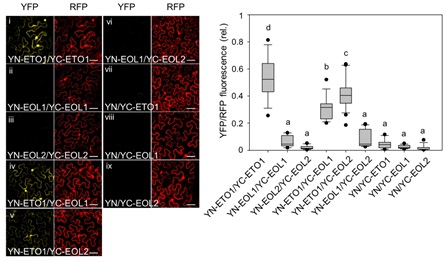
Figure. Ratiometric bimolecular fluorescence complementation (rBiFC) assay for protein interaction analysis of ETO1, EOL1, and EOL2.
Shin-Yuan Gu†, Wan-Sheng Lo†, Shaw-Jye Wu and Long-Chi Wang*.
ETHYLENE OVERPRODUCER1 (ETO1), ETO1-LIKE1 (EOL1) and EOL2 are members of the Broad-complex, Tramtrack, Bric-a-brac (BTB) protein family that collectively regulate type-2 1-aminocyclopropane -1-carboxylic acid synthase (ACS) activity. Despite ETO1 and EOL1/EOL2 encode structurally related proteins, genetic studies suggest that they do have redundant and distinct functions. To understand the mechanism in the ethylene biosynthesis, we studied the functional properties of the Arabidopsis ETO1 family. ETO1, EOL1 and EOL2 exhibit overlapping and distinct tissue-specific expression patterns. Nevertheless, neither EOL1 nor EOL2 can fully complement eto1 phenotype under the control of ETO1 promoter, which suggests differential functions of ETO1 and EOL1/EOL2. ETO1 forms homodimers and heterodimers with EOLs. Furthermore, CULLIN3 (CUL3) interacts preferentially with ETO1. The missense mutation in eto1‐5 generates a substitution of phenylalanine with an isoleucine in ETO1F466I that impairs its dimerization. This study provides a mechanistic view of such a regulation, which is dependent on the ability of ETO1 to form homodimers and dimerize with EOL1 and EOL2 to incorporate type‐2 ACS into the CUL3-dependent RING E3 ubiquitin ligase complex for protein ubiquitination followed by degradation via the 26S proteasome. This study reveals that protein-protein interactions of ETO1 and EOL1/EOL2 are crucial for their biological function in ethylene biosynthesis.